Cracking Chordoma's Conundrum: Immune Checkpoints Provide a Potential Modality
- PMID: 40386066
- PMCID: PMC12080586
- DOI: 10.7150/ijms.109721
Cracking Chordoma's Conundrum: Immune Checkpoints Provide a Potential Modality
Abstract
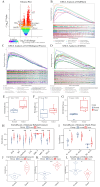
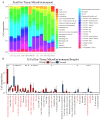
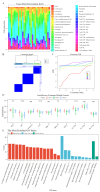
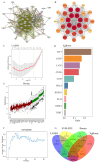
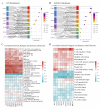
Objectives: Chordoma, a rare malignant tumor, is notably resistant to conventional treatments including chemotherapy, radiotherapy, and targeted approaches. Immunotherapy, successful in treating other cancer types, presents a promising avenue. However, the immune microenvironment of chordoma is poorly understood, highlighting the need to investigate immune checkpoints and their potential as therapeutic targets in this context. Methods: We performed an integrated analysis of chordoma using public datasets (GSE224776, GSE56183, GSE239531) and our RNA-seq data (11 samples). Differential expression analysis (limma), gene set enrichment analysis (GSEA, clusterProfiler), immune cell infiltration assessment (ESTIMATE, immunedeconv), weighted gene co-expression network analysis (WGCNA), consensus clustering, and machine learning were employed to identify key immune-related gene modules, immunogenic subtypes, and central immune regulators. Results: Hierarchical clustering and principal component analysis segregated chordoma from control samples post quality control. Differential expression analysis identified 2825 upregulated and 1693 downregulated genes, with significant upregulation of immune checkpoints, including PD-1 and CTLA-4. GSEA highlighted enhanced immune-related processes, particularly inflammatory responses, antigen presentation, and immune cell activation. Immune cell deconvolution demonstrated selective enrichment of memory T cells and macrophages, alongside downregulation of neutrophils and decreased effector cell scores. Consensus clustering identified a highly immunogenic chordoma subtype (Cluster 1), and WGCNA and machine learning converged on CCR7 as a central immune regulator, with core T cell-associated genes correlating with immune cell distribution patterns. Conclusion: This study characterizes the chordoma immune landscape, highlighting elevated immune checkpoints, distinct immunogenic subtypes, and a T cell-centered regulatory network. These findings support immune checkpoint inhibitors and other immunotherapies as promising treatments.
Keywords: chordoma; differential expression; immune checkpoint inhibitors; immune checkpoints.
© The author(s).
Conflict of interest statement
Competing interests: The authors have declared that no competing interest exists.
Figures

Similar articles
-
Identification of fatty acid metabolism-related molecular subtype biomarkers and their correlation with immune checkpoints in cutaneous melanoma.Front Immunol. 2022 Nov 18;13:967277. doi: 10.3389/fimmu.2022.967277. eCollection 2022. Front Immunol. 2022. PMID: 36466837 Free PMC article.
-
CXCR2P1 enhances the response of gastric cancer to PD-1 inhibitors through increasing the immune infiltration of tumors.Front Immunol. 2025 Mar 19;16:1545605. doi: 10.3389/fimmu.2025.1545605. eCollection 2025. Front Immunol. 2025. PMID: 40176817 Free PMC article.
-
Advancing personalized, predictive, and preventive medicine in bladder cancer: a multi-omics and machine learning approach for novel prognostic modeling, immune profiling, and therapeutic target discovery.Front Immunol. 2025 Apr 22;16:1572034. doi: 10.3389/fimmu.2025.1572034. eCollection 2025. Front Immunol. 2025. PMID: 40330458 Free PMC article.
-
Cancer immunotherapy resistance based on immune checkpoints inhibitors: Targets, biomarkers, and remedies.Drug Resist Updat. 2020 Dec;53:100718. doi: 10.1016/j.drup.2020.100718. Epub 2020 Jul 15. Drug Resist Updat. 2020. PMID: 32736034 Review.
-
Regulatory mechanisms of immune checkpoints PD-L1 and CTLA-4 in cancer.J Exp Clin Cancer Res. 2021 Jun 4;40(1):184. doi: 10.1186/s13046-021-01987-7. J Exp Clin Cancer Res. 2021. PMID: 34088360 Free PMC article. Review.
References
-
- Chambers KJ, Lin DT, Meier J, Remenschneider A, Herr M, Gray ST. Incidence and survival patterns of cranial chordoma in the United States. Laryngoscope. 2014;124:1097–102. - PubMed
-
- Stiller CA, Trama A, Serraino D, Rossi S, Navarro C, Chirlaque MD. et al. Descriptive epidemiology of sarcomas in Europe: report from the RARECARE project. Eur J Cancer. 2013;49:684–95. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

