Distribution of PAX1 and ZNF582 Hypermethylation in the Oral Exfoliated Cells of Oral Squamous Cell Carcinoma
- PMID: 40386310
- PMCID: PMC12084697
- DOI: 10.1177/11795549251335172
Distribution of PAX1 and ZNF582 Hypermethylation in the Oral Exfoliated Cells of Oral Squamous Cell Carcinoma
Abstract
Background: The DNA methylation statuses of PAX1 and ZNF582 show great promise as biomarkers for the detection of oral squamous cell carcinoma (OSCC). This study aims to investigate the distribution of PAX1 or ZNF582 methylation in the exfoliated oral epithelial cells (OECs) of OSCC.
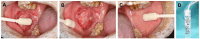
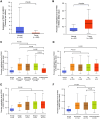
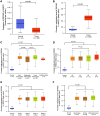
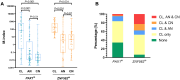
Methods: Methylation data from 528 tumors and 50 adjacent nontumor tissues were acquired from The Cancer Genome Atlas and analyzed using UALCAN database. Sixty-one OSCC cases from Peking University School and Hospital of Stomatology were included in this study and the exfoliated OECs collected by oral swabs were collected from the cancerous lesion (CL), adjacent normal (AN), and contralateral normal (CN) sites. The methylation levels of these 2 genes in different sites were evaluated.
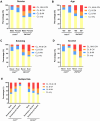
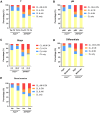
Results: PAX1 and ZNF582 were both hypermethylated in OSCC compared with nontumor sites but showed different methylation patterns within the oral environment. Generally, a CL-centric methylation pattern of PAX1 where methylation levels decrease gradually from CL through AN to CN was observed, suggesting a field cancerization effect. ZNF582 methylation levels are significantly higher at lesion sites compared with normal sites, but no significant difference is observed between AN and CN. Coexistence of ZNF582 methylation in CL and AN or CN sites was also observed in some patients with OSCC. Furthermore, ZNF582 methylation was more sensitive among patients with OSCC.
Conclusions: DNA methylation detection of PAX1 and ZNF582 in the exfoliated OECs is helpful for OSCC diagnosis. Hypermethylated PAX1 and ZNF582 show different methylation patterns in the oral cavity of patients with OSCC.
Keywords: DNA methylation; Oral squamous cell carcinoma; PAX1; ZNF582; field cancerization; oral swab; tumor suppressor gene.
© The Author(s) 2025.
Conflict of interest statement
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures
Similar articles
-
Hypermethylated PAX1 and ZNF582 genes in the tissue sample are associated with aggressive progression of oral squamous cell carcinoma.J Oral Pathol Med. 2020 Sep;49(8):751-760. doi: 10.1111/jop.13035. Epub 2020 Jun 3. J Oral Pathol Med. 2020. PMID: 32428271
-
Hypermethylated ZNF582 and PAX1 genes in oral scrapings collected from cancer-adjacent normal oral mucosal sites are associated with aggressive progression and poor prognosis of oral cancer.Oral Oncol. 2017 Dec;75:169-177. doi: 10.1016/j.oraloncology.2017.11.013. Epub 2017 Nov 17. Oral Oncol. 2017. PMID: 29224816
-
Evaluation of DNA methylation in matched oral swab and tissue specimens from Chinese patients with oral squamous cell carcinoma.Int J Oral Maxillofac Surg. 2021 Jun;50(6):725-732. doi: 10.1016/j.ijom.2020.05.022. Epub 2020 Oct 3. Int J Oral Maxillofac Surg. 2021. PMID: 33023801
-
The promising role of PAX1 (aliases: HUP48, OFC2) gene methylation in cancer screening.Mol Genet Genomic Med. 2019 Mar;7(3):e506. doi: 10.1002/mgg3.506. Epub 2019 Jan 12. Mol Genet Genomic Med. 2019. PMID: 30636379 Free PMC article. Review.
-
Epigenetic Regulation in Oral Squamous Cell Carcinoma Microenvironment: A Comprehensive Review.Cancers (Basel). 2023 Nov 27;15(23):5600. doi: 10.3390/cancers15235600. Cancers (Basel). 2023. PMID: 38067304 Free PMC article. Review.
References
-
- National Comprehensive Cancer Network. NCCN clinical practice guidelines in oncology (NCCN Guidelines): head and neck cancers, 2025.
LinkOut - more resources
Full Text Sources

