This is a preprint.
IBDome: An integrated molecular, histopathological, and clinical atlas of inflammatory bowel diseases
- PMID: 40386420
- PMCID: PMC12083645
- DOI: 10.21203/rs.3.rs-6443303/v1
IBDome: An integrated molecular, histopathological, and clinical atlas of inflammatory bowel diseases
Abstract
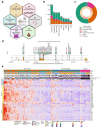
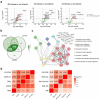
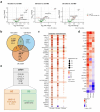
Multi-omic and multimodal datasets with detailed clinical annotations offer significant potential to advance our understanding of inflammatory bowel diseases (IBD), refine diagnostics, and enable personalized therapeutic strategies. In this multi-cohort study, we performed an extensive multi-omic and multimodal analysis of 1,002 clinically annotated patients with IBD and non-IBD controls, incorporating whole-exome and RNA sequencing of normal and inflamed gut tissues, serum proteomics, and histopathological assessments from images of H&E-stained tissue sections. Transcriptomic profiles of normal and inflamed tissues revealed distinct site-specific inflammatory signatures in Crohn's disease (CD) and ulcerative colitis (UC). Leveraging serum proteomics, we developed an inflammatory protein severity signature that reflects underlying intestinal molecular inflammation. Furthermore, foundation model-based deep learning accurately predicted histologic disease activity scores from images of H&Estained intestinal tissue sections, offering a robust tool for clinical evaluation. Our integrative analysis highlights the potential of combining multi-omics and advanced computational approaches to improve our understanding and management of IBD.
Conflict of interest statement
R.A. has served as a speaker, or consultant, or received research grants from AbbVie, Abivax, AlfaSigma, AstraZeneca, Bristol-Myers Squibb, CED Service GmbH, Celltrion Healthcare, Dr Falk Pharma, Galapagos, Johnson & Johnson, Eli Lilly, Materia Prima, MSD, Pfizer, and Takeda Pharma. J.N.K. declares consulting services for Bioptimus, France; Panakeia, UK; AstraZeneca, UK; and MultiplexDx, Slovakia. Furthermore, he holds shares in StratifAI, Germany, Synagen, Germany, Ignition Lab, Germany; has received an institutional research grant by GSK; and has received honoraria by AstraZeneca, Bayer, Daiichi Sankyo, Eisai, Janssen, Merck, MSD, BMS, Roche, Pfizer, and Fresenius. B.S. consulted for AbbVie, Abivax, Boehringer Ingelheim, Bristol Myers Squibb, Dr. Falk Pharma, Eli Lilly, Endpoint Health, Falk, Galapagos, Gilead, Janssen, Landos, Lilly, Materia Prima, PredictImmune, Pfizer, and Takeda; received speaker fees from AbbVie, AlfaSigma, BMS, CED Service GmbH, Dr. Falk Pharma, Eli Lilly, MSD, Ferring, Galapagos, Janssen, Pfizer, and Takeda; and received grant support from Pfizer (all the money went to an institutional account at Charité). All other authors declare no competing interests.
Figures


References
-
- Le Berre C., Ananthakrishnan A. N., Danese S., Singh S. & Peyrin-Biroulet L. Ulcerative Colitis and Crohn’s Disease Have Similar Burden and Goals for Treatment. Clin. Gastroenterol. Hepatol. 18, 14–23 (2020). - PubMed
-
- Wilson J. C., Furlano R. I., Jick S. S. & Meier C. R. Inflammatory Bowel Disease and the Risk of Autoimmune Diseases. J. Crohns Colitis 10, 186–193 (2016). - PubMed
-
- Dolinger M., Torres J. & Vermeire S. Crohn’s disease. Lancet Lond. Engl. 403, 1177–1191 (2024). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

