Ty retrotransposon element based multiple integration toolkit for Saccharomyces cerevisiae
- PMID: 40386439
- PMCID: PMC12083897
- DOI: 10.1016/j.synbio.2025.04.011
Ty retrotransposon element based multiple integration toolkit for Saccharomyces cerevisiae
Abstract
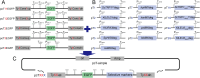
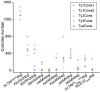
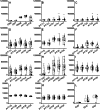
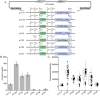
Extra-high-level overexpression of single or multiple specific proteins by integrating specific genes in the genome is vital to achieve the stable and efficient production of target proteins and metabolites in S. cerevisiae. Five families of Ty elements in the genome of S. cerevisiae CEN.PK2-1D, which could have dozens to hundreds of copies, have been employed to achieve massive gene expression. By engineering nine selective markers, six of them (TRP1, LEU2, URA3, HIS5, natMX and hphMX) achieve stably high copy integration (>15 copies) at Ty sites. Fluorescence proteins and taxifolin biosynthesis pathway genes were overexpressed to verify the toolkit. The titer of protein phiYFP in the multiple integration strain reached 1.6 g/L (268.1 mg/g DCW), and its fluorescence intensity was 3.3 times higher than that in the episomal overexpression strain. For taxifolin biosynthesis, 14 genes were integrated into three different Ty sites using three selective markers from the toolkit, resulting in 277.6 mg/L taxifolin accumulation from glucose.
Keywords: Gene overexpression; High copy number integration; Metabolic engineering; Protein production; Retrotransposons.
© 2025 The Authors.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials

