T7 RNA polymerase-guided base editor for accelerated continuous evolution in Bacillus subtilis
- PMID: 40386441
- PMCID: PMC12083895
- DOI: 10.1016/j.synbio.2025.04.010
T7 RNA polymerase-guided base editor for accelerated continuous evolution in Bacillus subtilis
Abstract
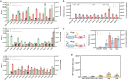
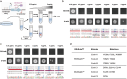
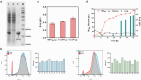
Targeted in vivo hypermutation mediated by base deaminase-T7 RNA polymerase (T7 RNAP) fusions promotes genetic diversification and accelerates continuous directed evolution. Due to the lack of a T7RNAP expression regulation system and functionally compatible linker for fusion protein expression, T7RNAP-guided continuous evolution has not been established in Bacillus subtilis, which limited long gene fragment continuous evolution targeted on genome. Here, we developed BS-MutaT7 system, which introduced mutations into specific genomic regions by leveraging chimeric fusions of base deaminases with T7RNAP in B. subtilis. We selected seven different sources of adenosine and cytosine deaminases, 14 fusion protein linkers to be fused with T7RNAP, constructing four libraries with the size of 5000, where deaminases were fused at either the N- or C-terminus of T7RNAP. Based on the efficiency of binding to T7 promoter and high mutagenesis activity, two optimal chimeric mutators, BS-MutaT7A (TadA8e-Linker0-T7RNAP) and BS-MutaT7C (PmCDA1-(GGGGS)3-T7RNAP co-expressed with UGI) were identified. The target mutation rates reached 1.2 × 10-5 per base per generation (s.p.b.) and 5.8 × 10-5 s.p.b., representing 7000-fold and 37,000-fold increases over the genomic mutation rate, respectively. Both exhibited high processivity, maintaining mutation rates of 5.8 × 10-6 s.p.b. and 2.9 × 10-5 s.p.b. within a 5 kb DNA region. Notably, BS-MutaT7C exhibited superior mutagenic activity, making it well-suited for applications requiring intensive and sustained genomic diversification. Application of BS-MutaT7 enabled a 16-fold increase in tigecycline resistance and enhanced β-lactoglobulin (β-Lg) expression by evolving the global transcriptional regulator codY, achieving a β-Lg titer of 3.92 g/L. These results highlight BS-MutaT7 as a powerful and versatile tool for genome-scale continuous evolution in B. subtilis.
Keywords: BS-MutaT7 system; Bacillus subtilis; Continuous directed evolution; T7 RNA polymerase.
© 2025 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
LinkOut - more resources
Full Text Sources

