Trio Whole Exome Sequencing in Chinese Childhood-Onset Lupus Reveals Novel Candidate Genes
- PMID: 40386946
- PMCID: PMC7617808
- DOI: 10.1002/art.43243
Trio Whole Exome Sequencing in Chinese Childhood-Onset Lupus Reveals Novel Candidate Genes
Abstract
Objective: Systemic lupus erythematosus (SLE) is an autoimmune disease in which rare and common gene variants contribute to pathogenesis. Severe sporadic disease in children is often explained by "de novo" variants that can be uncovered by trio sequencing.
Methods: Whole-exome sequencing was performed in 50 Chinese trios with childhood-onset SLE (cSLE). Rare coding variants in SLE-associated genes and all de novo variants were investigated. Gene pathway and expression analysis and interferon-β (IFNβ) luciferase assays were used to predict contribution to disease.
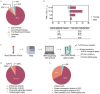
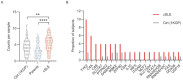
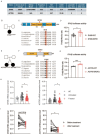
Results: Each proband carried at least one rare variant in an SLE-associated gene, with a median of six per child. At least two probands had monogenic disease, and one-third of probands carried novel or rare variants in genes well accepted to cause monogenic SLE: ACP5, C3, C4A, C4B, DNASE1, IFIH1, NRAS, RNASEH2B, RNASEH2C, and SAMHD1. Probands carried a median of one de novo, rare, coding variant. Intriguingly, although only two de novo variants occurred in genes previously associated with SLE, 12 of the 50 genes were enriched in the top 20 SLE-related pathways and were highly expressed in age-associated B cells and plasma B cells. These genes represent promising candidate lupus genes. Two de novo variants occurring in genes not previously linked to SLE or autoimmunity, DHX8 and ACTR5, enhanced type I IFN signaling.
Conclusion: This study highlights the abundance of lupus-relevant rare gene variants in cSLE, supports frequent contribution of de novo variants to disease, and identifies genes that may constitute novel therapeutic targets of relevance to Chinese patients.
© 2025 American College of Rheumatology.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

