Proliferating Microglia Exhibit Unique Transcriptional and Functional Alterations in Alzheimer's Disease
- PMID: 40387894
- PMCID: PMC12140498
- DOI: 10.1080/17590914.2025.2506406
Proliferating Microglia Exhibit Unique Transcriptional and Functional Alterations in Alzheimer's Disease
Abstract
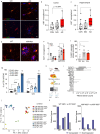
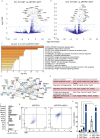
Proliferation of microglia represents a physiological process, which is accelerated in several neurodegenerative disorders including Alzheimer disease (AD). The effect of such neurodegeneration-associated microglial proliferation on function and disease progression remains unclear. Here, we show that proliferation results in profound alterations of cellular function by providing evidence that newly proliferated microglia show impaired beta-amyloid clearance in vivo. Through sorting of proliferating microglia of APP/PS1 mice and subsequent transcriptome analysis, we define unique proliferation-associated transcriptomic signatures that change with age and beta-amyloid accumulation and are characterized by enrichment of immune system-related pathways. Of note, we identify the DEAD-Box Helicase 3 X-Linked (DDX3X) as a key molecule to modulate microglia activation and cytokine secretion and it is expressed in the AD brain. Together, these results argue for a novel concept by which phenotypic and functional microglial changes occur longitudinally as a response to accelerated proliferation in a neurodegenerative environment.
Keywords: Alzheimer’s disease; inflammasome; microglia; proliferation; transcriptome.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

Similar articles
-
Amyloid precursor protein and presenilin-1 knock-in immunodeficient mice exhibit intraneuronal Aβ pathology, microgliosis, and extensive neuronal loss.Alzheimers Dement. 2025 Apr;21(4):e70084. doi: 10.1002/alz.70084. Alzheimers Dement. 2025. PMID: 40195277 Free PMC article.
-
Subtle Alterations in Hippocampal Neuronal Activity Coincide With Early Sex-Specific Differences in Amyloidosis and Microglia in a Pre-Symptomatic Mouse Model of Alzheimer-Like Pathology.Glia. 2025 Aug;73(8):1727-1745. doi: 10.1002/glia.70029. Epub 2025 Apr 30. Glia. 2025. PMID: 40304030
-
DNA hypomethylation promotes the expression of CASPASE-4 which exacerbates inflammation and amyloid-β deposition in Alzheimer's disease.Alzheimers Res Ther. 2024 Feb 8;16(1):29. doi: 10.1186/s13195-024-01390-2. Alzheimers Res Ther. 2024. PMID: 38326859 Free PMC article.
-
Microglial activation states and their implications for Alzheimer's Disease.J Prev Alzheimers Dis. 2025 Jan;12(1):100013. doi: 10.1016/j.tjpad.2024.100013. Epub 2025 Jan 1. J Prev Alzheimers Dis. 2025. PMID: 39800461 Free PMC article. Review.
-
CX3CL1 Pathway as a Molecular Target for Treatment Strategies in Alzheimer's Disease.Int J Mol Sci. 2023 May 4;24(9):8230. doi: 10.3390/ijms24098230. Int J Mol Sci. 2023. PMID: 37175935 Free PMC article. Review.
References
-
- Askew, K., Li, K., Olmos-Alonso, A., Garcia-Moreno, F., Liang, Y., Richardson, P., Tipton, T., Chapman, M. A., Riecken, K., Beccari, S., Sierra, A., Molnár, Z., Cragg, M. S., Garaschuk, O., Perry, V. H., & Gomez-Nicola, D. (2017). Coupled proliferation and apoptosis maintain the rapid turnover of microglia in the adult brain. Cell Reports, 18(2), 391–405. 10.1016/j.celrep.2016.12.041 - DOI - PMC - PubMed
-
- Bachiller, S., Jiménez-Ferrer, I., Paulus, A., Yang, Y., Swanberg, M., Deierborg, T., & Boza-Serrano, A. (2018). Microglia in neurological diseases: A road map to brain-disease dependent-inflammatory response. Frontiers in Cellular Neuroscience, 12, 488. 10.3389/fncel.2018.00488 - DOI - PMC - PubMed
-
- Chiu, I. M., Morimoto, E. T., Goodarzi, H., Liao, J. T., O’Keeffe, S., Phatnani, H. P., Muratet, M., Carroll, M. C., Levy, S., Tavazoie, S., Myers, R. M., & Maniatis, T. (2013). A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Reports, 4(2), 385–401. 10.1016/j.celrep.2013.06.018 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
