Whole-Blood RNA Sequencing Profiling of Patients With Rheumatoid Arthritis Treated With Tofacitinib
- PMID: 40388487
- PMCID: PMC12239517
- DOI: 10.1002/acr2.11761
Whole-Blood RNA Sequencing Profiling of Patients With Rheumatoid Arthritis Treated With Tofacitinib
Abstract
Objective: Patients with rheumatoid arthritis (RA) often fail to respond to therapies, including JAK inhibitors (JAKi), and treatment allocation is made via a trial-and-error strategy. A comprehensive analysis of responses to JAKi, including tofacitinib, by RNA sequencing (RNAseq) would allow the discovery of transcriptomic markers with a two-fold meaning: (1) an improved knowledge about the mechanisms of response to treatment (inference modeling) and (2) the definition of features that may be useful in treatment optimization and assignment (predictive modeling).
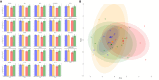
Methods: Thirty-three patients with active RA were treated with a tofacitinib dose of 5 mg twice a day for 24 weeks and evaluated for EULAR Disease Activity Score in 28 joints using the C-reactive protein level response. Whole-blood RNA was collected before and after treatment to perform RNAseq transcriptome analysis. Linear models were used to determine differentially expressed genes (DEGs) (1) at baseline according to clinical responses and (2) in the pre-post comparison after tofacitinib treatment and in relation to EULAR responses. The capability of DEGs to predict a successful treatment was tested via machine learning modeling after extensive internal validation.
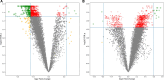
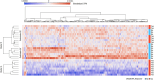
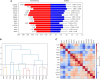
Results: Of 26 patients who completed the study (per-protocol analysis), 15 (57.7%) achieved good responses, and 7 (26.9%) and 4 (15.3%) had moderate and no responses, respectively. Overall, 273 baseline genes were significantly associated with the attainment of good responses, contributing to several pathways linked to the immune system or RA pathogenesis (eg, citrullination processes and the negative regulation of natural killer function). The expression of several molecules was reverted by tofacitinib when good responses were reached, including AKT3, GK5, KLF12, FCRL3, BIRC3, TSPOAP1, and P2RY10. Finally, we isolated 14 markers that singularly were capable of predicting the attainment of good responses, including, NKG2D, CD226, CLEC2D, and CD52.
Conclusion: Whole-blood transcriptome analysis of patients with RA treated with tofacitinib identified genes whose expression may be relevant in prognostication and understanding the mechanisms of responses to therapy.
© 2024 The Author(s). ACR Open Rheumatology published by Wiley Periodicals LLC on behalf of American College of Rheumatology.
Figures
References
-
- Smolen JS, Landewé RBM, Bijlsma JWJ, et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease‐modifying antirheumatic drugs: 2019 update. Ann Rheum Dis 2020;79(6):685–699. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

