Association between circulating inflammatory proteins and gout: A Mendelian randomization study
- PMID: 40388724
- PMCID: PMC12091660
- DOI: 10.1097/MD.0000000000042379
Association between circulating inflammatory proteins and gout: A Mendelian randomization study
Abstract
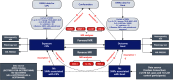
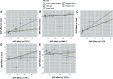
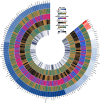
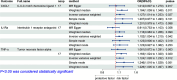
Clinical studies have consistently demonstrated that inflammation is a critical factor in the pathophysiology and progression of gout. This study aims to explore the causal relationship between CIPs and gout, utilizing MR in conjunction with meta-analyses. We utilized genetic data pertaining to gout from the GWAS which involved 3576 cases and 147,221 control participants. A total of 132 CIPs were extracted from the GWAS data to identify SNPs associated with gout. The primary analytical approach was the IVW method. Sensitivity analyses indicated no pleiotropy or heterogeneity. The IVW results revealed that several CIPs were associated with gout in European populations. The analysis results indicate that FGF-21, MMP-1, G-CSF, and IFN-γ are involved in the pathogenesis of gout, and gout may influence the expression of CXCL1, IL-1Ra, and TNF-α. Consequently, targeted research focusing on specific CIPs could provide a promising strategy for the treatment and prevention of gout, offering potential therapeutic targets for the underlying inflammatory mechanisms of the disease.
Keywords: Mendelian randomization; bidirectional; circulating inflammatory proteins; gout; meta-analysis.
Copyright © 2025 the Author(s). Published by Wolters Kluwer Health, Inc.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures



References
-
- Dehlin M, Jacobsson L, Roddy E. Global epidemiology of gout: prevalence, incidence, treatment patterns and risk factors. Nat Rev Rheumatol. 2020;16:380–90. - PubMed
-
- Hachicha M, Naccache PH, McColl SR. Inflammatory microcrystals differentially regulate the secretion of macrophage inflammatory protein 1 and interleukin 8 by human neutrophils: a possible mechanism of neutrophil recruitment to sites of inflammation in synovitis. J Exp Med. 1995;182:2019–25. - PMC - PubMed
-
- Migliorini P, Italiani P, Pratesi F, Puxeddu I, Boraschi D. The IL-1 family cytokines and receptors in autoimmune diseases. Autoimmun Rev. 2020;19:102617. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

