PERRC: Protease Engineering with Reactant Residence Time Control
- PMID: 40388903
- PMCID: PMC12301959
- DOI: 10.1021/acssynbio.5c00154
PERRC: Protease Engineering with Reactant Residence Time Control
Abstract
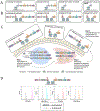
Proteases with engineered specificity hold great potential for targeted therapeutics, protein circuit construction, and biotechnology applications. However, many proteases exhibit broad substrate specificity, limiting their use in such applications. Engineering protease specificity remains challenging because evolving a protease to recognize a new substrate, without counterselecting against its native substrate, often results in high residual activity on the original substrate. To address this, we developed Protease Engineering with Reactant Residence Time Control (PERRC), a platform that exploits the correlation between endoplasmic reticulum (ER) retention sequence strength and ER residence time. PERRC allows precise control over the stringency of protease evolution by adjusting counterselection to selection substrate ratios. Using PERRC, we evolved an orthogonal tobacco etch virus protease variant, TEVESNp, that selectively cleaves a substrate (ENLYFES) that differs by only one amino acid from its parent sequence (ENLYFQS). TEVESNp exhibits a remarkable 65-fold preference for the evolved substrate, marking the first example of an engineered orthogonal protease driven by such a slight difference in substrate recognition. Furthermore, TEVESNp functions as a competent protease for constructing orthogonal protein circuits in bacteria, and molecular dynamics simulations analysis reveals subtle yet functionally significant active site rearrangements. PERRC is a modular dual-substrate display system that facilitates precise engineering of protease specificity.
Keywords: high-throughput screening; protease engineering; synthetic biology; tobacco etch virus (TEV) protease; yeast surface display.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
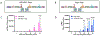
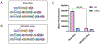
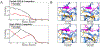
Update of
-
PERRC: Protease Engineering with Reactant Residence Time Control.bioRxiv [Preprint]. 2025 Mar 4:2025.03.02.641063. doi: 10.1101/2025.03.02.641063. bioRxiv. 2025. Update in: ACS Synth Biol. 2025 Jun 20;14(6):2241-2253. doi: 10.1021/acssynbio.5c00154. PMID: 40093119 Free PMC article. Updated. Preprint.
Similar articles
-
PERRC: Protease Engineering with Reactant Residence Time Control.bioRxiv [Preprint]. 2025 Mar 4:2025.03.02.641063. doi: 10.1101/2025.03.02.641063. bioRxiv. 2025. Update in: ACS Synth Biol. 2025 Jun 20;14(6):2241-2253. doi: 10.1021/acssynbio.5c00154. PMID: 40093119 Free PMC article. Updated. Preprint.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Sexual Harassment and Prevention Training.2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36508513 Free Books & Documents.
-
Interventions to prevent occupational noise-induced hearing loss.Cochrane Database Syst Rev. 2017 Jul 7;7(7):CD006396. doi: 10.1002/14651858.CD006396.pub4. Cochrane Database Syst Rev. 2017. PMID: 28685503 Free PMC article.
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

