Single-cell RNA-sequencing reveals early mitochondrial dysfunction unique to motor neurons shared across FUS- and TARDBP-ALS
- PMID: 40389397
- PMCID: PMC12089458
- DOI: 10.1038/s41467-025-59679-1
Single-cell RNA-sequencing reveals early mitochondrial dysfunction unique to motor neurons shared across FUS- and TARDBP-ALS
Abstract
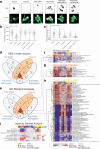
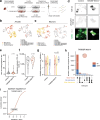
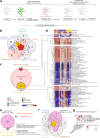
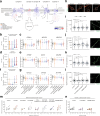
Mutations in FUS and TARDBP cause amyotrophic lateral sclerosis (ALS), but the precise mechanisms of selective motor neuron degeneration remain unresolved. To address if pathomechanisms are shared across mutations and related to either gain- or loss-of-function, we performed single-cell RNA sequencing across isogenic induced pluripotent stem cell-derived neuron types, harbouring FUS P525L, FUS R495X, TARDBP M337V mutations or FUS knockout. Transcriptional changes were far more pronounced in motor neurons than interneurons. About 20% of uniquely dysregulated motor neuron transcripts were shared across FUS mutations, half from gain-of-function. Most indicated mitochondrial impairments, with attenuated pathways shared with mutant TARDBP M337V as well as C9orf72-ALS patient motor neurons. Mitochondrial motility was impaired in ALS motor axons, even with nuclear localized FUS mutants, demonstrating shared toxic gain-of-function mechanisms across FUS- and TARDBP-ALS, uncoupled from protein mislocalization. These early mitochondrial dysfunctions unique to motor neurons may affect survival and represent therapeutic targets in ALS.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors have no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

