Transcriptional and epigenetic rewiring by the NUP98::KDM5A fusion oncoprotein directly activates CDK12
- PMID: 40389480
- PMCID: PMC12089343
- DOI: 10.1038/s41467-025-59930-9
Transcriptional and epigenetic rewiring by the NUP98::KDM5A fusion oncoprotein directly activates CDK12
Abstract
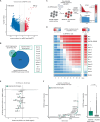
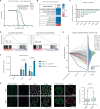
Nucleoporin 98 (NUP98) fusion oncoproteins are strong drivers of pediatric acute myeloid leukemia (AML) with poor prognosis. Here we show that NUP98 fusion-expressing AML harbors an epigenetic signature that is characterized by increased accessibility of hematopoietic stem cell genes and enrichment of activating histone marks. We employ an AML model for ligand-induced degradation of the NUP98::KDM5A fusion oncoprotein to identify epigenetic programs and transcriptional targets that are directly regulated by NUP98::KDM5A through CUT&Tag and nascent RNA-seq. Orthogonal genome-wide CRISPR/Cas9 screening identifies 12 direct NUP98::KDM5A target genes, which are essential for AML cell growth. Among these, we validate cyclin-dependent kinase 12 (CDK12) as a druggable vulnerability in NUP98::KDM5A-expressing AML. In line with its role in the transcription of DNA damage repair genes, small-molecule-mediated CDK12 inactivation causes increased DNA damage, leading to AML cell death. Altogether, we show that NUP98::KDM5A directly regulates a core set of essential target genes and reveal CDK12 as an actionable vulnerability in AML with oncogenic NUP98 fusions.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.Z. is a founder, shareholder, and scientific advisor of Quantro Therapeutics GmbH. J.Z. and the Zuber laboratory receive research support and funding from Boehringer Ingelheim. P.V. received consultancy honoraria from Pfizer, AOP Orphan, Delbert, Novartis, and Blueprint. The other authors declare no conflict of interest.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

