Tumour-derived microparticles obtained through microwave irradiation induce immunogenic cell death in lung adenocarcinoma
- PMID: 40389640
- PMCID: PMC12373502
- DOI: 10.1038/s41565-025-01922-3
Tumour-derived microparticles obtained through microwave irradiation induce immunogenic cell death in lung adenocarcinoma
Abstract
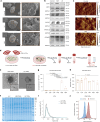
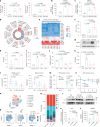
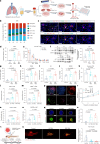
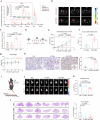
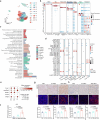
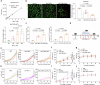
Tumour-derived microparticles (TMPs), extracellular vesicles traditionally obtained upon ultraviolet (UV) radiation of tumour cells, hold promise in tumour immunotherapies and vaccines and have demonstrated potential as drug delivery systems for tumour treatment. However, concerns remain regarding the limited efficacy and safety of UV-derived TMPs. Here we introduce a microwave (MW)-assisted method for preparing TMPs, termed MW-TMPs. Brief exposure of tumour cells to short-wavelength MW radiation promotes the release of TMPs showing superior in vivo antitumour activity and safety compared with UV-TMPs. MW-TMPs induce immunogenic cell death and reprogramme suppressive tumour immune microenvironments in different lung tumour models, enabling dual targeting of tumour cells by natural killer and T cells. We show that they can efficiently deliver methotrexate to tumours, synergistically boosting the efficacy of PD-L1 blockade. This MW-TMP development strategy is simpler, more efficient and safer than traditional UV-TMP methods.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures







References
-
- Prakash, A., Gates, T., Zhao, X., Wangmo, D. & Subramanian, S. Tumor-derived extracellular vesicles in the colorectal cancer immune environment and immunotherapy. Pharmacol. Ther.241, 108332 (2023). - PubMed
MeSH terms
Grants and funding
- 82330003/National Natural Science Foundation of China (National Science Foundation of China)
- 82270110/National Natural Science Foundation of China (National Science Foundation of China)
- 82200112/National Natural Science Foundation of China (National Science Foundation of China)
- 82100112/National Natural Science Foundation of China (National Science Foundation of China)
- 2022YFF1203300/Chinese Ministry of Science and Technology | Department of S and T for Social Development (Department of S&T for Social Development)
- 2023YFC0872500/Chinese Ministry of Science and Technology | Department of S and T for Social Development (Department of S&T for Social Development)
- 2022BCA016/Department of Science and Technology, Hubei Provincial People's Government (Hubei Provincial Department of Science and Technology)
- 2023AFB1073/Natural Science Foundation of Hubei Province (Hubei Provincial Natural Science Foundation)
LinkOut - more resources
Full Text Sources
Medical
Research Materials

