ANXA1 promotes intrahepatic cholangiocarcinoma proliferation and growth by regulating glutamine metabolism through GOT1 stabilization
- PMID: 40390008
- PMCID: PMC12087091
- DOI: 10.1186/s13046-025-03400-z
ANXA1 promotes intrahepatic cholangiocarcinoma proliferation and growth by regulating glutamine metabolism through GOT1 stabilization
Abstract
Background: Intrahepatic cholangiocarcinoma (ICC) is a malignant tumor with a poor prognosis, marked by a postoperative recurrence rate of 50-60% and a 5-year survival rate of 8-30%. Abnormal tumor metabolism, particularly, amino acid metabolism, plays a key role in malignant progression. However, the molecular mechanisms linking amino acid metabolism to ICC progression remain unclear.
Methods: Bioinformatics was used to identity the key amino acid metabolism related gene in ICC, qRT-PCR, western blotting and immunohistochemical (IHC) were used to detect the expression of ANXA1 in normal tissues or ICC tissues and cells at mRNA and protein levels. The effects of ANXA1 on the proliferation ability of ICC in vitro and in vivo were investigated using CCK8, cloning formation experiment, EdU, IHC, nude mice subcutaneous tumorigenesis model. Immunoprecipitation, mass spectrometry analysis, protein ubiquitin level detection test, immunofluorescence co-localization, and redox stress metabolite detection test were used to explore the metabolism-related regulatory mechanism of ANXA1.
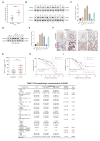
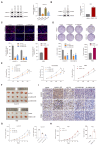
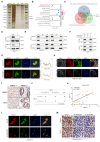
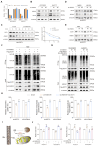
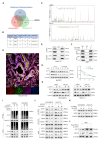
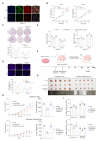
Results: we employed bioinformatics analysis to classify ICC into metabolic subgroups with distinct prognoses and identified the associated biomarker Annexin A1(ANXA1), whose high expression is correlated with poor prognosis and promotes ICC development. Mass spectrometry analysis revealed that ANXA1 interacts with the key enzyme in glutamine metabolism, glutamic-oxaloacetic transaminase 1(GOT1). Through in vitro and in vivo experiments, overexpressed ANXA1 stabilizes GOT1 by recruiting the deubiquitinase USP5. This stabilization enhances glutamine uptake, as well as the production of aspartate and glutamate, which in turn reduces oxidative stress, thereby promoting tumor cell growth. Moreover, knockdown of ANXA1 combined with glutamine uptake inhibition significantly suppressed ICC cell proliferation and Inhibited subcutaneous tumor formation and growth.
Conclusions: These results suggest that the ANXA1/USP5/GOT1 axis promotes glutamine metabolism and ICC proliferation and growth. Inhibiting ANXA1 alongside glutamine uptake inhibition offers a promising strategy for treating ICC.
Keywords: ANXA1; GOT1; Glutamine metabolism; ICC; Oxidative stress.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study was supported by the ethics committee of the first affiliated hospital of Guizhou Medical University and the Animal Care and Treatment Committee of Guizhou Medical University. Consent for publication: All the authors agreed to publish the study in the journal. Competing interests: The authors declare no competing interests. relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Annexin A1: A new immunohistological marker of cholangiocarcinoma.World J Gastroenterol. 2013 Apr 28;19(16):2456-65. doi: 10.3748/wjg.v19.i16.2456. World J Gastroenterol. 2013. PMID: 23674846 Free PMC article.
-
Zinc Finger Protein 263 Augments Autophagy and Promotes Intrahepatic Cholangiocarcinoma Proliferation.Mol Carcinog. 2025 Feb;64(2):317-328. doi: 10.1002/mc.23847. Epub 2024 Nov 18. Mol Carcinog. 2025. PMID: 39555729
-
Autocrine parathyroid hormone-like hormone promotes intrahepatic cholangiocarcinoma cell proliferation via increased ERK/JNK-ATF2-cyclinD1 signaling.J Transl Med. 2017 Nov 25;15(1):238. doi: 10.1186/s12967-017-1342-1. J Transl Med. 2017. PMID: 29178939 Free PMC article.
-
Glutamic oxaloacetic transaminase 1 as a potential target in human cancer.Eur J Pharmacol. 2022 Feb 15;917:174754. doi: 10.1016/j.ejphar.2022.174754. Epub 2022 Jan 8. Eur J Pharmacol. 2022. PMID: 35007521 Review.
-
The role of Tripartite motif containing 59 (TRIM59) in the proliferation and prognosis of intrahepatic cholangiocarcinoma.Pathol Res Pract. 2022 Aug;236:153989. doi: 10.1016/j.prp.2022.153989. Epub 2022 Jun 17. Pathol Res Pract. 2022. PMID: 35753134 Review.
References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. Jan. 2022;72(1):7–33. 10.3322/caac.21708. - PubMed
-
- Khan SA, Tavolari S, Brandi G. Cholangiocarcinoma: epidemiology and risk factors. Liver Int. May 2019;39(1):19–31. 10.1111/liv.14095. - PubMed
-
- Forner A, Vidili G, Rengo M, Bujanda L, Ponz-Sarvisé M, Lamarca A. Clinical presentation, diagnosis and staging of cholangiocarcinoma. Liver Int. May 2019;39(1):98–107. 10.1111/liv.14086. - PubMed
-
- El-Diwany R, Pawlik TM, Ejaz A. Intrahepatic Cholangiocarcinoma, Surg Oncol Clin N Am, vol. 28, no. 4, pp. 587–599, Oct. 2019, 10.1016/j.soc.2019.06.002 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

