Dual-responsive synthetic gene circuit for dynamic biologic drug delivery via inflammatory and circadian signaling pathways
- PMID: 40390078
- PMCID: PMC12090394
- DOI: 10.1186/s13036-025-00519-7
Dual-responsive synthetic gene circuit for dynamic biologic drug delivery via inflammatory and circadian signaling pathways
Abstract
Background: Engineered cells provide versatile tools for precise, tunable drug delivery, especially when synthetic stimulus-responsive gene circuits are incorporated. In many complex disease conditions, endogenous pathologic signals such as inflammation can vary dynamically over different time scales. For example, in autoimmune conditions such as rheumatoid arthritis or juvenile idiopathic arthritis, local (joint) and systemic inflammatory signals fluctuate daily, peaking in the early morning, but can also persist over long periods of time, triggering flare-ups that can last weeks to months. However, treatment with disease-modifying anti-rheumatic drugs is typically provided at continuous high doses, regardless of disease activity and without consideration for levels of inflammatory signals. In previous studies, we have developed cell-based drug delivery systems that can automatically address the different scales of flares using either chronogenetic circuits (i.e., clock gene-responsive elements) that can be tuned for optimal drug delivery to dampen circadian variations in inflammatory levels or inflammation-responsive circuits (i.e., NF-κB-sensitive elements) that can respond to sustained arthritis flares on demand with proportional synthesis of drug. The goal of this study was to develop a novel dual-responsive synthetic gene circuit that responds to both circadian and inflammatory inputs using OR-gate logic for both daily timed therapeutic output and enhanced therapeutic output during chronic inflammatory conditions.
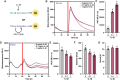
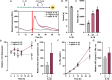
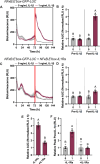
Results: We developed a synthetic gene circuit driven by tandem inflammatory NF-κB and circadian E'-box response elements. When engineered into induced pluripotent stem cells that were chondrogenically differentiated, the gene circuit demonstrated basal-level circadian output with enhanced stimulus-responsive output during an inflammatory challenge shown by bioluminescence monitoring. Similarly, this system exhibited enhanced therapeutic levels of biologic drug interleukin-1 receptor antagonist (IL-1Ra) during an inflammatory challenge in differentiated cartilage pellets. This dual-responsive therapeutic gene circuit mitigated both the inflammatory response as measured by bioluminescence reporter output and tissue-level degradation during conditions mimicking an arthritic flare.
Conclusions: The dual-responsive synthetic gene circuit developed herein responds to input cues from two key homeostatic transcriptional networks, enabling dynamic and tunable output. This proof-of-concept approach has the potential to match drug delivery to disease activity for optimal outcomes that addresses the complex environment of inflammatory arthritis.
Keywords: Arthritis; Cartilage; Cell engineering; Chronogenetic; Circadian rhythms; Gene circuits; Synthetic biology.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: FG is an employee and shareholder in Cytex Therapeutics, Inc. FG has filed intellectual property on topics related to the content of this study (US Patent App. 18/284,487, 2024). The authors declare no other competing interests.
Figures


Update of
-
Dual-responsive synthetic gene circuit for dynamic biologic drug delivery via inflammatory and circadian signaling pathways.bioRxiv [Preprint]. 2025 Mar 20:2025.03.20.644403. doi: 10.1101/2025.03.20.644403. bioRxiv. 2025. Update in: J Biol Eng. 2025 May 19;19(1):47. doi: 10.1186/s13036-025-00519-7. PMID: 40166199 Free PMC article. Updated. Preprint.
Similar articles
-
Dual-responsive synthetic gene circuit for dynamic biologic drug delivery via inflammatory and circadian signaling pathways.bioRxiv [Preprint]. 2025 Mar 20:2025.03.20.644403. doi: 10.1101/2025.03.20.644403. bioRxiv. 2025. Update in: J Biol Eng. 2025 May 19;19(1):47. doi: 10.1186/s13036-025-00519-7. PMID: 40166199 Free PMC article. Updated. Preprint.
-
Programmable chronogenetic gene circuits for self-regulated circadian delivery of biologic drugs.J Control Release. 2025 Sep 10;385:113959. doi: 10.1016/j.jconrel.2025.113959. Epub 2025 Jun 18. J Control Release. 2025. PMID: 40541742
-
Programmable chronogenetic gene circuits for self-regulated circadian delivery of biologic drugs.bioRxiv [Preprint]. 2025 Mar 15:2025.03.14.643274. doi: 10.1101/2025.03.14.643274. bioRxiv. 2025. Update in: J Control Release. 2025 Sep 10;385:113959. doi: 10.1016/j.jconrel.2025.113959. PMID: 40161636 Free PMC article. Updated. Preprint.
-
Advanced application of stimuli-responsive drug delivery system for inflammatory arthritis treatment.Mater Today Bio. 2022 Feb 21;14:100223. doi: 10.1016/j.mtbio.2022.100223. eCollection 2022 Mar. Mater Today Bio. 2022. PMID: 35243298 Free PMC article. Review.
-
Gene therapy for rheumatoid arthritis. Lessons from animal models, including studies on interleukin-4, interleukin-10, and interleukin-1 receptor antagonist as potential disease modulators.Rheum Dis Clin North Am. 2002 Feb;28(1):127-49. doi: 10.1016/s0889-857x(03)00073-5. Rheum Dis Clin North Am. 2002. PMID: 11840694 Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

