Enhancing infectious intestinal disease diagnosis through metagenomic and metatranscriptomic sequencing of 1000 human diarrhoeal samples
- PMID: 40390150
- PMCID: PMC12090668
- DOI: 10.1186/s13073-025-01478-w
Enhancing infectious intestinal disease diagnosis through metagenomic and metatranscriptomic sequencing of 1000 human diarrhoeal samples
Abstract
Background: Current surveillance of diarrhoeal disease is hindered by limitations of traditional diagnostic approaches, which often fail to identify the causative organism, particularly for novel or hard-to-culture bacterial pathogens. Sequencing nucleic acids directly from stool can overcome such constraints, but such approaches need to reliably detect pathogens identifiable by conventional methods.
Methods: As part of the INTEGRATE study, we analysed stool microbiomes from 1067 patients with gastroenteritis symptoms using direct sequencing, and compared findings with standard diagnostic techniques (culture, immunoassay, microscopy, and single-target PCR) and molecular assays (Luminex xTAG GPP) for detection of bacterial and viral pathogens in the UK.
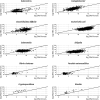
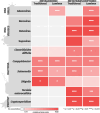
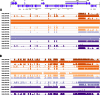
Results: We found strong positive correlations between metatranscriptomic reads and traditional diagnostics for six out of 15 pathogens. The metatranscriptomic data were highly correlated with the Luminex assay for eight out of 14 pathogens. In contrast, metagenomic sequencing only showed a strong positive correlation with traditional diagnostics for three of 15 pathogens, and with Luminex for four of 14 pathogens. Compared with metagenomics, metatranscriptomics had increased sensitivity of detection for four pathogens, while metagenomics was more effective for detecting five pathogens. Metatranscriptomics gave near-complete transcriptome coverage for Human mastadenovirus F and detected Cryptosporidium via identification of Cryptosporidium parvum virus (CSpV1). A comprehensive transcriptomic profile of Salmonella enterica serovar Enteritidis was recovered from the stool of a patient with a laboratory-confirmed Salmonella infection. Furthermore, comparison of RNA/DNA ratios between pathogen-positive and pathogen-negative samples demonstrated that metatranscriptomics can distinguish pathogen-positive/negative samples and provide insights into pathogen biology. Higher RNA/DNA ratios were observed in samples that tested positive via gold-standard diagnostics.
Conclusions: This study highlights the power of directly sequencing nucleic acids from human samples to augment gastrointestinal pathogen surveillance and clinical diagnostics. Metatranscriptomics was most effective for identifying a wide range of pathogens and showed superior sensitivity. We propose that metatranscriptomics should be considered for future diagnosis and surveillance of gastrointestinal pathogens. We assembled a rich data resource of paired metagenomic and metatranscriptomic datasets, direct from patient stool samples, and have made these data publicly available to enhance the understanding of pathogens associated with infectious intestinal diseases.
Keywords: Culture-independent; Diagnostics; Genomics; Metagenome; Metatranscriptome; Microbiome; Pathogens.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Members of the public with symptoms of acute gastroenteritis, including a case definition of vomiting and diarrhoea, who sought health advice from general practices in the RCGP RSC NMN were invited to submit a stool sample for microbiological examination. Their consent for this procedure was sought because normal care would not necessarily entail stool sampling for most patients unless their symptoms were severe or had persisted for a long time. The North West - Greater Manchester East Research Ethics Committee (REC reference: 15/NW/0233) and NHS Health Research Authority (HRA) Confidential Advisory Group (CAG) (CAG reference: 15/CAG/0131) granted a favourable ethics opinion for the INTEGRATE project. Approval was also granted by NHS Research Management and Governance Committees (including Royal Liverpool and Broadgreen University Hospital Trust, Lancashire Teaching Hospitals NHS Foundation Trust, Central Manchester University Hospitals NHS Foundation Trust, and the University of Liverpool Sponsor), the Lancaster University Faculty of Health and Medicine Ethics Committee, and the University of Liverpool Ethics Sub-Committees. An Information Governance Toolkit (IGT) from the Department of Health hosted by the Health and Social Care Information Centre (HSCIC) was also completed for the project, and all project research staff obtained Honorary NHS contracts, research passports, and letters of access, as necessary. We confirm that this research conforms to the principles of the Helsinki Declaration. Consent for publication: The publication was approved by the National Institute for Health and Care Research on 29 th March 2023. Competing interests: M.I.G. has received research grants from GSK and Merck, and has provided expert advice to GSK. M.I.G. has been an employee of GSK since January 2023, although the work presented here was completed prior to this date. The remaining authors declare that they have no competing interests.
Figures

References
-
- Food Standards Agency. Foodborne disease estimates for the United Kingdom in 2018. 2020. Available from: https://webarchive.nationalarchives.gov.uk/ukgwa/20200803160512/https://.... Accessed 9 Sep 2024.
-
- UK Government. Public Health England, NHS. UK Standards for Microbiology Investigations - Gastroenteritis; 2020. Available from: https://assets.publishing.service.gov.uk/government/uploads/system/uploa.... Accessed 9 Sep 2024.
-
- Kanagarajah S, Waldram A, Dolan G, Jenkins C, Ashton PM, Carrion Martin AI, et al. Whole genome sequencing reveals an outbreak of Salmonella Enteritidis associated with reptile feeder mice in the United Kingdom, 2012–2015. Food Microbiol. 2018;71:32–8. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

