Design and Evaluation of Andrographolide Analogues as SARS-CoV-2 Main Protease Inhibitors: Molecular Modeling and in vitro Studies
- PMID: 40391180
- PMCID: PMC12087591
- DOI: 10.2147/DDDT.S514193
Design and Evaluation of Andrographolide Analogues as SARS-CoV-2 Main Protease Inhibitors: Molecular Modeling and in vitro Studies
Abstract
Background: The COVID-19 pandemic, caused by SARS-CoV-2, highlights the urgent need for novel antiviral agents targeting key viral proteins. The main protease (Mpro) is a crucial enzyme for viral replication, making it an attractive drug target. Andrographolide, a natural compound with known antiviral properties, serves as a promising scaffold for inhibitor development.
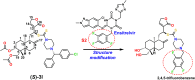
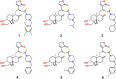
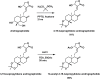
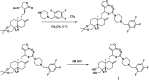
Objective: This study aimed to design, synthesize, and evaluate C-12 dithiocarbamate andrographolide analogues as potential SARS-CoV-2 Mpro inhibitors using computational and experimental approaches.
Methods: A structure-based drug design approach was employed to design andrographolide derivatives. Molecular dynamics simulations were conducted to assess binding interactions and stability. The hit compound was synthesized and evaluated using an enzyme inhibition assay against SARS-CoV-2 Mpro. Cytotoxicity was assessed in HepG2, HaCaT, and HEK293T cells to determine safety profiles.
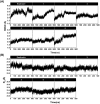
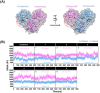
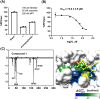
Results: Among the designed compounds, compound 1, incorporating a 2,4,5-trifluorobenzene moiety, exhibited the strongest binding affinity and stable interactions with key Mpro residues (H41, M49 and M165). Enzyme inhibition assay confirmed ~70% inhibition at 100 µM, with moderate to low cytotoxicity (CC50 values comparable to andrographolide).
Conclusion: Compound 1 represents a promising non-covalent SARS-CoV-2 Mpro inhibitor. Further structural optimization is necessary to enhance potency, selectivity, and safety for therapeutic applications.
Keywords: COVID-19; MD simulations; SARS-CoV-2 main protease; andrographolide analogues; enzyme-based assay.
© 2025 Suriya et al.
Conflict of interest statement
The authors declare no conflicts of interest in this work.
Figures


Similar articles
-
Repurposing FDA-approved drugs for COVID-19: targeting the main protease through multi-phase in silico approach.Antivir Ther. 2024 Dec;29(6):13596535241305536. doi: 10.1177/13596535241305536. Antivir Ther. 2024. PMID: 39639531
-
Design, synthesis, and biological evaluation of dithiocarbamate derivatives as SARS-CoV-2 Mpro inhibitors.Bioorg Med Chem Lett. 2024 Dec 1;114:130011. doi: 10.1016/j.bmcl.2024.130011. Epub 2024 Oct 30. Bioorg Med Chem Lett. 2024. PMID: 39486486
-
Discovery of Severe Acute Respiratory Syndrome Coronavirus 2 Main Protease Inhibitors through Rational Design of Novel Fluorinated 1,3,4-oxadiazole Amide Derivatives: An In-Silico Study.Chem Biodivers. 2025 Jun;22(6):e202403179. doi: 10.1002/cbdv.202403179. Epub 2025 Feb 14. Chem Biodivers. 2025. PMID: 39853882
-
Non-peptidic inhibitors targeting SARS-CoV-2 main protease: A review.Bioorg Chem. 2024 Jun;147:107380. doi: 10.1016/j.bioorg.2024.107380. Epub 2024 Apr 16. Bioorg Chem. 2024. PMID: 38636432 Review.
-
An Updated Review on SARS-CoV-2 Main Proteinase (MPro): Protein Structure and Small-Molecule Inhibitors.Curr Top Med Chem. 2021;21(6):442-460. doi: 10.2174/1568026620666201207095117. Curr Top Med Chem. 2021. PMID: 33292134 Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

