TNFRSF12A expression in stomach adenocarcinoma and its preliminary role in predicting immunotherapy response
- PMID: 40391219
- PMCID: PMC12086169
- DOI: 10.3389/fimmu.2025.1578068
TNFRSF12A expression in stomach adenocarcinoma and its preliminary role in predicting immunotherapy response
Abstract
Background: TNFRSF12A is abnormally expressed in various malignancies, especially in stomach adenocarcinoma (STAD), which is related to tumor invasiveness and prognosis of patients. This study examined the expression pattern of TNFRSF12A in STAD and predicted immunotherapy response.
Methods: Data were derived from The Cancer Gene Atlas (TCGA), Gene Expression Omnibus (GEO), and Gene Expression Profiling Interactive Analysis (GEPIA) to analyze the expression pattern of TNFRSF12A in pan-cancer and STAD, as well as its correlation with clinical features. Biological pathways involved in TNFRSF12A were analyzed by "clusterProfiler" package. Immune cell infiltration was evaluated by "GSVA" and "CIBERSORT" packages. Immunotherapy response was assessed by TIDE score and tumor mutation burden (TMB) level. Expression level of TNFRSF12A in the single cell of STAD was analyzed by scRNA-seq. Finally, in vitro test detected the mRNA expression of TNFRSF12A in STAD cells, Wound healing and Transwell assays were performed to measure the capabilities of STAD cell to migrate and invade.
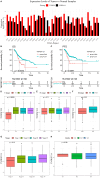
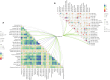
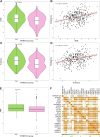
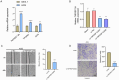
Results: TNFRSF12A was highly expressed in STAD. However, TNFRSF12A expression did not shown significant difference in relation to clinical features. TNFRSF12A exhibited notably positive correlation with many carcinogenic signaling pathways and immune cells infiltration such as T cells and macrophages. High TNFRSF12A expression group showed a higher TIDE score, Exclusion score, and TMB level than the low TNFRSF12A expression group, which indicated that STAD patients with high TNFRSF12A expression responded more poorly to immunotherapy. TNFRSF12A showed a positive relation with most of immune checkpoint genes. By scRNA-seq analysis, TNFRSF12A was chiefly expressed in Fibroblasts and Mast cells of STAD. Further, in vitro assays verified the high expression of TNFRSF12A in STAD cells, and the migration and invasion capabilities of STAD cells were notably suppressed by TNFRSF12A silencing (p<0.05).
Conclusion: The present study not only reveals the potential of TNFRSF12A as a therapeutic target for STAD, but also explores its great potential in STAD immunotherapy. This finding opens up a new way of thinking for the personalized treatment of STAD.
Keywords: TNFRSF12A; immunotherapy; single-cell RNA sequencing; stomach adenocarcinoma; tumor microenvironment.
Copyright © 2025 Sun, Zhang, Wan, Yang, Yao, Yang, Liu and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Fatima S, Song Y, Zhang Z, Fu Y, Zhao R, Malik K, et al. . Exploring the pharmacological mechanisms of P-hydroxylcinnamaldehyde for treating gastric cancer: A pharmacological perspective with experimental confirmation. Curr Mol Pharmacol. (2024) 17:e18761429322420. doi: 10.2174/0118761429322420241112051105 - DOI - PubMed
-
- Chandarana CV, Mithani NT, Singh DV, Kikani UB. Vibrational spectrophotometry: A comprehensive review on the diagnosis of gastric and liver cancer. Curr Pharm Anal. (2024) 20:453–65. doi: 10.2174/0115734129322567240821052326 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

