Fighting RNA viruses with a gold nanoparticle Cas13d gene-editing armor
- PMID: 40391300
- PMCID: PMC12088821
- DOI: 10.1016/j.omtn.2025.102540
Fighting RNA viruses with a gold nanoparticle Cas13d gene-editing armor
Abstract
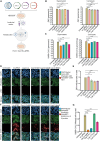
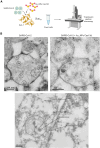
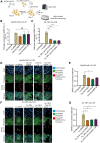
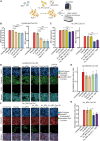
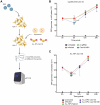
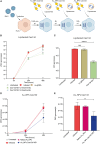
A novel Cas13d-based gene-editing approach has been developed to target viral RNAs in infected cells, reducing the replication of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and Zika virus (ZIKV) by up to 90% compared with controls. Despite its potential, the use of Cas13d as an antiviral faces several challenges that limit its effectiveness before reaching target cells. This study presents a proof-of-concept strategy for constructing Cas13d with gold nanoparticles (Au_NPs) to destroy SARS-CoV-2 and ZIKV genomes into cells. The Au_NPs Cas13d complexes were administered to Huh-7 cells infected with either virus, in single or multiple doses. The study demonstrated that Au_NPs Cas13d cuts target RNAs with comparable efficiency as lipofected ribonucleoprotein (RNP). Additionally, we found that Au_NPs Cas13d can spontaneously enter cells by endocytosis or diffusion, before the first 4 h of treatment. Au_NPs Cas13d co-localized with SARS-CoV-2 virions in early endosomes and reduced SARS-CoV-2 replication after a single administration, unlike RNPs, which showed no antiviral activity. However, Au_NPs Cas13d was less efficient at reducing ZIKV replication compared with lipofected Cas13d-RNPs, likely due to different intracellular localization. These results suggest that Au_NPs can be adapted as a new antiviral strategy, highlighting an innovative delivery method of Cas13d against viruses without the need for transfecting, providing a new gene-editing-based approach against emerging RNA viruses.
Keywords: Au_NPs Cas13d; Cas13d; MT: RNA/DNA Editing; RNA viruses; SARS-CoV-2; Zika virus; emerging viruses; gold nanoparticles.
© 2025 The Authors.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Evaluation of the effect of RNA secondary structure on Cas13d-mediated target RNA cleavage.Mol Ther Nucleic Acids. 2024 Jul 20;35(3):102278. doi: 10.1016/j.omtn.2024.102278. eCollection 2024 Sep 10. Mol Ther Nucleic Acids. 2024. PMID: 39220269 Free PMC article.
-
CRISPR-Cas13d effectively targets SARS-CoV-2 variants, including Delta and Omicron, and inhibits viral infection.MedComm (2020). 2023 Jan 31;4(1):e208. doi: 10.1002/mco2.208. eCollection 2023 Feb. MedComm (2020). 2023. PMID: 36744219 Free PMC article.
-
Efficient CRISPR-Cas13d-Based Antiviral Strategy to Combat SARS-CoV-2.Viruses. 2023 Mar 6;15(3):686. doi: 10.3390/v15030686. Viruses. 2023. PMID: 36992394 Free PMC article.
-
Manipulation of genes could inhibit SARS-CoV-2 infection that causes COVID-19 pandemics.Exp Biol Med (Maywood). 2021 Jul;246(14):1643-1649. doi: 10.1177/15353702211008106. Epub 2021 Apr 25. Exp Biol Med (Maywood). 2021. PMID: 33899542 Free PMC article. Review.
-
Cas13d: A New Molecular Scissor for Transcriptome Engineering.Front Cell Dev Biol. 2022 Mar 31;10:866800. doi: 10.3389/fcell.2022.866800. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35433685 Free PMC article. Review.
Cited by
-
Gold nanoparticle-based delivery of Cas13d for targeted RNA virus defense.Mol Ther Nucleic Acids. 2025 Aug 8;36(3):102652. doi: 10.1016/j.omtn.2025.102652. eCollection 2025 Sep 9. Mol Ther Nucleic Acids. 2025. PMID: 40822030 Free PMC article. No abstract available.
References
-
- Fiolet T., Kherabi Y., MacDonald C.J., Ghosn J., Peiffer-Smadja N. Comparing COVID-19 vaccines for their characteristics, efficacy and effectiveness against SARS-CoV-2 and variants of concern: a narrative review. Clin. Microbiol. Infect. 2022;28:202–221. doi: 10.1016/j.cmi.2021.10.005. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

