A single-base mutation in promoter of CsTPR enhances the negative regulation on mechanical-related leaf drooping in tea plants
- PMID: 40391387
- PMCID: PMC12087448
- DOI: 10.1093/hr/uhaf098
A single-base mutation in promoter of CsTPR enhances the negative regulation on mechanical-related leaf drooping in tea plants
Abstract
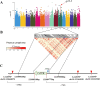
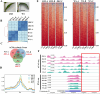
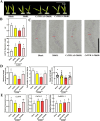
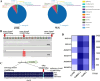
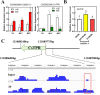
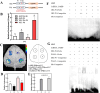
Mechanical harvesting in the tea industry has become increasingly essential due to its advantages in increasing productivity and reducing labor costs. Leaf droopiness caused a high rate of broken leaves, hindering the mechanized harvesting quality. However, the underlying mechanisms remain unclear. We herein identified a quantitative trait locus, designated as q10.3, along with three lead single nucleotide polymorphisms (SNPs) located near a TPR gene (TETRATRICOPEPTIDE REPEAT), named CsTPR, through performing a genome-wide association study (GWAS) on 130 tea accessions. Integrated analysis of RNA-seq and ATAC-seq confirmed CsTPR as a droopiness-associated candidate gene at the transcriptional level. CsTPR was then proved to negatively regulate brassinosteroid-induced droopiness by using the CsTPR-silencing tea plant. Whole-genome sequencing (WGS) combined with genome walking further indicated that a single-base mutation (T-A) in the promoter of CsTPR. ChIP-seq revealed that this mutation occurred within the binding site, E-box, of CsBES1.2 on the CsTPR promoter. Notably, CsBES1.2 bound the E-box of CsTPR promoter to repress the expression of CsTPR, as demonstrated by chromatin immunoprecipitation quantitative polymerase chain reaction (ChIP-qPCR), electrophoretic mobility shift assays (EMSA), and transient assays. The single-base mutation strengthened the inhibitory effect of CsBES1.2 on the expression of CsTPR via enhancing the binding affinity to the E-box. Altogether, our findings suggest that CsTPR negatively regulates droopiness in tea plants under the transcriptional repression of CsBES1.2 and that a single-base mutation within E-box amplifies the suppression of CsBES1.2 on the expression of CsTPR.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nanjing Agricultural University.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Similar articles
-
CsEXL3 regulate mechanical harvest-related droopy leaves under the transcriptional activation of CsBES1.2 in tea plant.Hortic Res. 2024 Mar 7;11(5):uhae074. doi: 10.1093/hr/uhae074. eCollection 2024 May. Hortic Res. 2024. PMID: 38738211 Free PMC article.
-
Genome-wide association study of leaf-related traits in tea plant in Guizhou based on genotyping-by-sequencing.BMC Plant Biol. 2023 Apr 12;23(1):196. doi: 10.1186/s12870-023-04192-0. BMC Plant Biol. 2023. PMID: 37046207 Free PMC article.
-
Genome-Wide Identification, Characterization, and Expression Analysis of BES1 Family Genes in 'Tieguanyin' Tea Under Abiotic Stress.Plants (Basel). 2025 Feb 5;14(3):473. doi: 10.3390/plants14030473. Plants (Basel). 2025. PMID: 39943035 Free PMC article.
-
Genome-Wide Identification, Structural, and Gene Expression Analysis of BRI1-EMS-Suppressor 1 Transcription Factor Family in Cucumis sativus.Front Genet. 2020 Oct 6;11:583996. doi: 10.3389/fgene.2020.583996. eCollection 2020. Front Genet. 2020. PMID: 33133168 Free PMC article.
-
Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond.Cell Cycle. 2014;13(18):2847-52. doi: 10.4161/15384101.2014.949201. Cell Cycle. 2014. PMID: 25486472 Free PMC article. Review.
Cited by
-
Transcriptome sequencing revealed the regulation of stem internode length associated with mechanical harvesting in three tea varieties.Front Plant Sci. 2025 Jun 30;16:1626915. doi: 10.3389/fpls.2025.1626915. eCollection 2025. Front Plant Sci. 2025. PMID: 40661767 Free PMC article.
References
-
- Jiang C, Moon D, Ma J. et al. Characteristics of non-volatile metabolites in fresh shoots from tea plant (Camellia sinensis) and its closely related species and varieties. Beverage Plant Res. 2022;2:1–15
-
- Kumar KR, Dashora K, Kumar S. et al. A review of drying technology in tea sector of industrial , non-conventional and renewable energy based drying systems. Appl Therm Eng. 2023;224:1, 120118–25
-
- Yu Y, Li W, Liu Y. et al. A Zea genus-specific micropeptide controls kernel dehydration in maize. Cell. 2024;188:1–16 - PubMed
LinkOut - more resources
Full Text Sources

