A Feedback Loop Between Fatty Acid Metabolism and Epigenetics in Clear Cell Renal Carcinoma
- PMID: 40391655
- PMCID: PMC12302539
- DOI: 10.1002/advs.202504532
A Feedback Loop Between Fatty Acid Metabolism and Epigenetics in Clear Cell Renal Carcinoma
Abstract
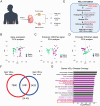
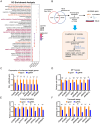
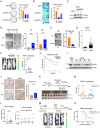
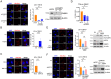
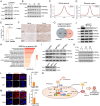
Lipid storage and epigenetic dysregulation are key features for clear cell renal carcinoma (ccRCC). However, the interplay between fatty acid metabolism and epigenetics in ccRCC remains to be further demonstrated. Here, the landscape of active enhancers is profiled in paired ccRCC samples and identifies 10171 gain variant enhancer loci (VELs) in the tumor tissues. Experimental validation reveals the enhancers targeting FABP5, FABP6, LPCAT1, MET, SEMA5B, SH3GL1, SNX33, and RHBDF2 are oncogenic. Further studies in organoids and animal models prove FABP5 as an oncogene. HIF-2α and ZNF692 transcription factors regulate FABP5 expression through directly binding to its promoter and enhancer. FABP5 is essential for the lipid droplet formation driven by HIFs and critical for H3K27ac and enhancer activity in ccRCC cells. Thus, the study has identified potential targets for drug design and diagnosis and discovered the function of a feedback loop between epigenetics and lipid metabolism regulated by FABP5 in ccRCC.
Keywords: FABP5; H3K27ac; ccRCC; enhancer; lipid droplet.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- a) Morgan M. A., Shilatifard A., Genes Dev. 2015, 29, 238; - PMC - PubMed
- b) Brookes E., Shi Y., Annu. Rev. Genet. 2014, 48, 237; - PubMed
- c) Sharma S., Kelly T. K., Jones P. A., Carcinogenesis 2010, 31, 27; - PMC - PubMed
- d) Gu M., Ren B., Fang Y., Ren J., Liu X., Wang X., Zhou F., Xiao R., Luo X., You L., Zhao Y., MedComm 2024, 5, 495; - PMC - PubMed
- e) Zou Y., Yang A., Chen B., Deng X., Xie J., Dai D., Zhang J., Tang H., Wu T., Zhou Z., Xie X., Wang J., Drug Resistance Updates 2024, 77, 101126. - PubMed
-
- Shlyueva D., Stampfel G., Stark A., Nat. Rev. Genet. 2014, 15, 272. - PubMed
MeSH terms
Substances
Grants and funding
- 2023YFA0913400/National Key R&D Program of China
- 2023YFC2507405/National Key R&D Program of China
- 32470620/National Natural Science Foundation of China
- 3217050383/National Natural Science Foundation of China
- 2020BCB007/Fundamental Research Funds for the Central Universities, Key Research and Development Program of Hubei Province
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
