Linear dicentric chromosomes in bacterial natural isolates reveal common constraints for replicon fusion
- PMID: 40391973
- PMCID: PMC12153318
- DOI: 10.1128/mbio.01046-25
Linear dicentric chromosomes in bacterial natural isolates reveal common constraints for replicon fusion
Abstract
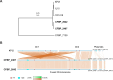
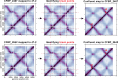
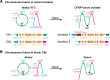
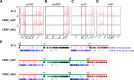
Multipartite bacterial genome organization can confer advantages, including coordinated gene regulation and faster genome replication, but is challenging to maintain. Agrobacterium tumefaciens lineages often contain a circular chromosome (Ch1), a linear chromosome (Ch2), and multiple plasmids. We previously observed that in some stocks of the C58 lab model, Ch1 and Ch2 were fused into a linear dicentric chromosome. Here we analyzed Agrobacterium natural isolates from the French Collection for Plant-Associated Bacteria and identified two strains distinct from C58 with fused chromosomes. Chromosome conformation capture identified integration junctions that were different from the C58 fusion strain. Genome-wide DNA replication profiling showed that both replication origins remained active. Transposon sequencing revealed that partitioning systems of both chromosome centromeres were essential. Importantly, the site-specific recombinase XerCD is required for the survival of the strains containing the fusion chromosome. Our findings show that replicon fusion occurs in natural environments and that balanced replication arm sizes and proper resolution systems enable the survival of such strains.
Importance: Most bacterial genomes are monopartite with a single, circular chromosome. However, some species, like Agrobacterium tumefaciens, carry multiple chromosomes. Emergence of multipartite genomes is often related to adaptation to specific niches, including pathogenesis or symbiosis. Multipartite genomes confer certain advantages; however, maintaining this complex structure can present significant challenges. We previously reported a laboratory-propagated lineage of A. tumefaciens strain C58 in which the circular and linear chromosomes fused to form a single dicentric chromosome. Here we discovered two geographically separated environmental isolates of A. tumefaciens containing fused chromosomes with integration junctions different from the C58 fusion chromosome, revealing the constraints and diversification of this process. We found that balanced replication arm sizes and the repurposing of multimer resolution systems enable the survival and stable maintenance of dicentric chromosomes. These findings reveal how multipartite genomes function across different bacterial species and the role of genomic plasticity in bacterial genetic diversification.
Keywords: Agrobacterium tumefaciens; Hi-C; chromosome fusion; multipartite genome; natural isolates.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Update of
-
Linear dicentric chromosomes in bacterial natural isolates reveal common constraints for replicon fusion.bioRxiv [Preprint]. 2025 Feb 24:2025.02.23.639760. doi: 10.1101/2025.02.23.639760. bioRxiv. 2025. Update in: mBio. 2025 Jun 11;16(6):e0104625. doi: 10.1128/mbio.01046-25. PMID: 40060587 Free PMC article. Updated. Preprint.
Similar articles
-
Linear dicentric chromosomes in bacterial natural isolates reveal common constraints for replicon fusion.bioRxiv [Preprint]. 2025 Feb 24:2025.02.23.639760. doi: 10.1101/2025.02.23.639760. bioRxiv. 2025. Update in: mBio. 2025 Jun 11;16(6):e0104625. doi: 10.1128/mbio.01046-25. PMID: 40060587 Free PMC article. Updated. Preprint.
-
A dicentric bacterial chromosome requires XerC/D site-specific recombinases for resolution.Curr Biol. 2022 Aug 22;32(16):3609-3618.e7. doi: 10.1016/j.cub.2022.06.050. Epub 2022 Jul 6. Curr Biol. 2022. PMID: 35797999 Free PMC article.
-
Conformation and dynamic interactions of the multipartite genome in Agrobacterium tumefaciens.Proc Natl Acad Sci U S A. 2022 Feb 8;119(6):e2115854119. doi: 10.1073/pnas.2115854119. Proc Natl Acad Sci U S A. 2022. PMID: 35101983 Free PMC article.
-
Integration of genes into the chromosome of Agrobacterium tumefaciens C58.Methods Mol Biol. 2006;343:55-66. doi: 10.1385/1-59745-130-4:55. Methods Mol Biol. 2006. PMID: 16988333 Review.
-
Divided genomes: negotiating the cell cycle in prokaryotes with multiple chromosomes.Mol Microbiol. 2005 Jun;56(5):1129-38. doi: 10.1111/j.1365-2958.2005.04622.x. Mol Microbiol. 2005. PMID: 15882408 Review.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

