Neuron-like macrophage differentiation via the APOE-TREM2 axis contributes to chronic pain in nasopharyngeal carcinoma
- PMID: 40392335
- PMCID: PMC12092482
- DOI: 10.1007/s10565-025-10035-5
Neuron-like macrophage differentiation via the APOE-TREM2 axis contributes to chronic pain in nasopharyngeal carcinoma
Abstract
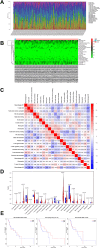
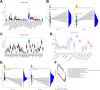
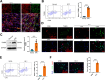
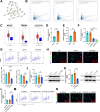
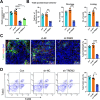
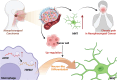
Chronic pain is a prevalent and debilitating symptom in patients with nasopharyngeal carcinoma (NPC). Fresh insights indicate that tumor-associated macrophages (TAMs) within the tumor microenvironment (TME) may undergo neuron-like differentiation, potentially contributing to pain mechanisms. By examining the apolipoprotein E (APOE) together with the triggering receptor expressed on myeloid cells 2 (TREM2), this study aims to clarify their joint function in modulating differentiation and how this interplay might be implicated in chronic pain associated with NPC. Through comprehensive analysis using TCGA-NPC transcriptomic datasets and single-cell RNA sequencing (scRNA-seq), we assessed the molecular landscapes of both NPC-affected and healthy nasopharyngeal tissues. Differential gene expression and immune cell profiling identified macrophages as key players in the inflammatory response. Single-cell sequencing revealed a distinct subpopulation of neuron-like macrophages expressing neurogenesis-related genes. Macrophage-to-neuron-like cell transformation in response to NPC cells was examined through in vitro co-culture systems, highlighting the involvement of the APOE-TREM2 regulatory pathway. In vivo studies involved macrophage depletion and TREM2 knockdown in mouse models to evaluate the impact on chronic pain development. Infiltrating macrophages were significantly more abundant in NPC samples, with many exhibiting neuron-like features that were positively linked to high levels of WNT5 A expression. In vitro, NPC cells induced macrophage differentiation into neuron-like cells, a process regulated by TREM2 and APOE. TREM2 knockdown in macrophages resulted in a reduction of chronic pain behaviors in mouse models, highlighting the contribution of the APOE-TREM2 Axis to NPC-associated chronic pain. Our findings demonstrate that NPC cells promote macrophage reprogramming through the APOE-TREM2 Axis, leading to neuron-like differentiation and contributing to chronic pain in NPC patients. Targeting this pathway may offer novel therapeutic strategies for managing chronic pain in NPC.
Keywords: Chronic pain; Nasopharyngeal carcinoma; Neuron-like macrophages; Single-cell sequencing; TCGA; TREM2.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study adheres to the Helsinki Declaration and has obtained informed consent from the patients, as well as approval from the ethics committee of Shengjing Hospital of China Medical University for conducting medical research. All animal experiments conducted in this research institute have received approval from the Animal Ethics Committee of Shengjing Hospital of China Medical University and adhere to local principles for the management and utilization of experimental animals. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
TREM2 knockdown improves the therapeutic effect of PD-1 blockade in hepatocellular carcinoma.Biochem Biophys Res Commun. 2022 Dec 25;636(Pt 1):140-146. doi: 10.1016/j.bbrc.2022.10.079. Epub 2022 Oct 25. Biochem Biophys Res Commun. 2022. PMID: 36332476
-
KMO-driven metabolic reconfiguration and its impact on immune cell infiltration in nasopharyngeal carcinoma: a new avenue for immunotherapy.Cancer Immunol Immunother. 2025 Feb 1;74(3):75. doi: 10.1007/s00262-024-03928-7. Cancer Immunol Immunother. 2025. PMID: 39891699 Free PMC article.
-
Huoxue Tongluo tablet enhances atherosclerosis efferocytosis by promoting the differentiation of Trem2+ macrophages via PPARγ signaling pathway.Phytomedicine. 2025 May;140:156579. doi: 10.1016/j.phymed.2025.156579. Epub 2025 Feb 26. Phytomedicine. 2025. PMID: 40068297
-
The Role of APOE and TREM2 in Alzheimer's Disease-Current Understanding and Perspectives.Int J Mol Sci. 2018 Dec 26;20(1):81. doi: 10.3390/ijms20010081. Int J Mol Sci. 2018. PMID: 30587772 Free PMC article. Review.
-
New Classification of Macrophages in Plaques: a Revolution.Curr Atheroscler Rep. 2020 Jun 18;22(8):31. doi: 10.1007/s11883-020-00850-y. Curr Atheroscler Rep. 2020. PMID: 32556603 Review.
References
-
- Chang ET, Ye W, Zeng Y-X, Adami H-O. The Evolving Epidemiology of Nasopharyngeal Carcinoma. Cancer Epidemiol Biomarkers Prev. American Association for Cancer Research (AACR); 2021 [cited 2025 Apr 18]. p. 1035–47. Available from: 10.1158/1055-9965.epi-20-1702 - PubMed
-
- Chen S, Peng J, Sherchan P, Ma Y, Xiang S, Yan F, et al. TREM2 activation attenuates neuroinflammation and neuronal apoptosis via PI3K/Akt pathway after intracerebral hemorrhage in mice. J Neuroinflammation. Springer Science and Business Media LLC; 2020 [cited 2025 Apr 18]. Available from: 10.1186/s12974-020-01853-x - PMC - PubMed
-
- Cohen SP, Vase L, Hooten WM. Chronic pain: an update on burden, best practices, and new advances. Lancet. Elsevier BV; 2021 [cited 2025 Apr 18]. p. 2082–97. Available from: 10.1016/s0140-6736(21)00393-7 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

