Microbiome and pediatric leukemia, diabetes, and allergies: Systematic review and meta-analysis
- PMID: 40392825
- PMCID: PMC12091780
- DOI: 10.1371/journal.pone.0324167
Microbiome and pediatric leukemia, diabetes, and allergies: Systematic review and meta-analysis
Abstract
Background: Despite the different pathologies and genetic susceptibilities of childhood ALL, T1DM and allergies, these conditions share epidemiological risk factors related to timing of infectious exposures and acquisition of the gut microbiome in infancy. We have assessed whether lower microbiome diversity (Shannon Index) and shared genus/species profiles are associated with pediatric ALL, allergies, and T1DM.
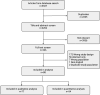
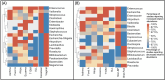
Methods and findings: Literature search was performed using PubMed, Embase, Cochrane, and Web of Science databases. Case-control, meta-analyses, and cohort studies were considered for inclusion. Inclusion criteria: (i) subjects age 1-18 years at diagnosis, (ii) reports effect of microbiome measured prior to/at time of diagnosis/first intervention (iii) outcome of ALL, allergies, asthma, or T1DM, (iv) English text. Exclusion criteria: (i) age < 1 or >18 years at diagnosis, (ii) Down Syndrome-associated ALL, (iii) non-English text, (iv) reviews, pre-print, or abstracts, (v) heavily biased studies. Abstract and full text screening were performed by two independent reviewers. Data extraction was performed by one reviewer following PRISMA guidelines. Data were pooled using a random-effects model. Eighty-eight studies were included in the analysis, with seventy-seven in the qualitative analysis and 54 in the meta-analysis. Cases were found to have lower alpha-diversity than controls in ALL (SMD:-0.78, 95%CI:-1.21, -0.34), T1DM (SMD:-1.26, 95%CI:-3.49, 0.96), eczema (SMD:-0.34, 95%CI:-0.56, -0.12), atopy (SMD:-0.06, 95%CI:-0.34, 0.22), asthma (SMD:-0.37, 95%CI:-1.16, 0.42), and food allergy (SMD:-0.11, 95%CI:-0.63, 0.41).
Conclusions: These results highlight similarities in the microbiome diversity and composition of children with ALL, T1DM, and allergies. This is compatible with a common risk factor related to immune priming in infancy and highlights the gut microbiome as a potentially modifiable risk factor and preventative strategy for these childhood diseases.
Copyright: © 2025 Gallant et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Gut microbiome-based interventions for the management of obesity in children and adolescents aged up to 19 years.Cochrane Database Syst Rev. 2025 Jul 10;7(7):CD015875. doi: 10.1002/14651858.CD015875. Cochrane Database Syst Rev. 2025. PMID: 40637175 Review.
-
Education support services for improving school engagement and academic performance of children and adolescents with a chronic health condition.Cochrane Database Syst Rev. 2023 Feb 8;2(2):CD011538. doi: 10.1002/14651858.CD011538.pub2. Cochrane Database Syst Rev. 2023. PMID: 36752365 Free PMC article.
-
Prognosis of adults and children following a first unprovoked seizure.Cochrane Database Syst Rev. 2023 Jan 23;1(1):CD013847. doi: 10.1002/14651858.CD013847.pub2. Cochrane Database Syst Rev. 2023. PMID: 36688481 Free PMC article.
-
Interventions for promoting habitual exercise in people living with and beyond cancer.Cochrane Database Syst Rev. 2018 Sep 19;9(9):CD010192. doi: 10.1002/14651858.CD010192.pub3. Cochrane Database Syst Rev. 2018. PMID: 30229557 Free PMC article.
-
Physical exercise training interventions for children and young adults during and after treatment for childhood cancer.Cochrane Database Syst Rev. 2016 Mar 31;3(3):CD008796. doi: 10.1002/14651858.CD008796.pub3. Cochrane Database Syst Rev. 2016. PMID: 27030386 Free PMC article.
References
-
- Tamburini S, Shen N, Wu H, Clemente J. The microbiome in early life: implications for health outcomes. Nat Med. 2016;22(7):713–22. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

