Prevalence and novel genetic characteristics of Cryptosporidium spp. in wild rodents in the northern foothills of the Dabie Mountains, southeast Henan Province, China
- PMID: 40392880
- PMCID: PMC12091789
- DOI: 10.1371/journal.pntd.0013117
Prevalence and novel genetic characteristics of Cryptosporidium spp. in wild rodents in the northern foothills of the Dabie Mountains, southeast Henan Province, China
Abstract
Background: Cryptosporidium spp. are prevalent zoonotic pathogens that affect both humans and animals. The pathogens are spread through feces and represent a major cause of diarrhea. As they are both abundant and widely distributed, wild rodents play a significant role in the transmission of Cryptosporidium spp. The Dabie Mountains in southeast Henan Province are rich in wildlife resources as well as various species of livestock. However, the epidemiological characteristics of Cryptosporidium spp. among local wild rodents remain poorly understood. Therefore, the infection rate and genetic characteristics of Cryptosporidium spp. in wild rodents within this region should be determined.
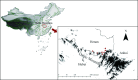
Methods: Between March 2023 and April 2024, a total of 267 wild rodents were captured in the northern foothills of the Dabie Mountains, and fecal samples were collected from their intestines for DNA extraction. Species identification of wild rodents was conducted using PCR amplification of the universal vertebrate cytochrome b (cytb) gene. Nested PCR was subsequently used to amplify the small subunit (SSU) rRNA, actin, heat shock protein 70 (HSP70), and 60 kDa glycoprotein (gp60) genes for the analysis of Cryptosporidium species, genotypes, and subtypes in the fecal samples.
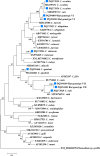
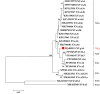
Results: The infection rate of Cryptosporidium spp. in wild rodents from the northern foothills of the Dabie Mountains was 21.3% (57/267). Seven species of wild rodents were identified, and the infection rates for Cryptosporidium spp. varied among host species. In particular, the infection rate was 21.4% (25/117) in Niviventer lotipes, 22.4% (22/98) in Apodemus agrarius, 17.2% (5/29) in Rattus nitidus, 22.2% (4/18) in Apodemus draco, and 33.3% (1/3) in Rattus tanezumi. The identification results indicated the presence of five Cryptosporidium species: Cryptosporidium apodemi (n = 12), C. ubiquitum (n = 11), C. viatorum (n = 7), C. ratti (n = 2), and C. occultus (n = 2). Moreover, two novel genotypes were identified: Cryptosporidium sp. rat genotype VI (n = 8) and Cryptosporidium sp. rat genotype VII (n = 15). Notably, a new subtype of C. viatorum designated as XVgA4 was discovered.
Conclusions: This study revealed the prevalence of Cryptosporidium spp. in wild rodents in the northern foothills of the Dabie Mountains and identified two novel Cryptosporidium genotypes, along with a new subtype, C. viatorum-XVgA4. The findings highlight the genetic diversity of Cryptosporidium spp., underscoring the increased risk of Cryptosporidium spp. transmission posed by local wild rodents population. It suggests that host-specific factors should be considered in epidemiological surveillance and control strategies of Cryptosporidium spp., which is of great significance for the prevention and control of Cryptosporidiosis.
Copyright: © 2025 Yang et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Feng K, Yang S, Xu Y, Wen L, Chen J, Zhang W, et al. Molecular characterization of Cryptosporidium spp., Giardia spp. and Enterocytozoon bieneusi in eleven wild rodent species in China: Common distribution, extensive genetic diversity and high zoonotic potential. One Health. 2024;18:100750. doi: 10.1016/j.onehlt.2024.100750 - DOI - PMC - PubMed
-
- Zhou S, Jiang Y, Cao J. Cryptosporidium infection in rodents in China. Chin J Parasitol Parasit Dis. 2024;42:512–20.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

