Spatial domain detection using contrastive self-supervised learning for spatial multi-omics technologies
- PMID: 40393810
- PMCID: PMC12212350
- DOI: 10.1101/gr.279380.124
Spatial domain detection using contrastive self-supervised learning for spatial multi-omics technologies
Abstract
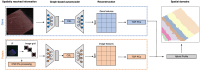
Recent advances in spatially resolved single-omic and multi-omics technologies have led to the emergence of computational tools to detect and predict spatial domains. Additionally, histological images and immunofluorescence (IF) staining of proteins and cell types provide multiple perspectives and a more complete understanding of tissue architecture. Here, we introduce Proust, a scalable tool to predict discrete domains using spatial multi-omics data by combining the low-dimensional representation of biological profiles based on graph-based contrastive self-supervised learning. Our scalable method integrates multiple data modalities, such as RNA, protein, and H&E images, and predicts spatial domains within tissue samples. Through the integration of multiple modalities, Proust consistently demonstrates enhanced accuracy in detecting spatial domains, as evidenced across various benchmark data sets and technological platforms.
© 2025 Yao et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



Update of
-
Spatial domain detection using contrastive self-supervised learning for spatial multi-omics technologies.bioRxiv [Preprint]. 2024 Feb 4:2024.02.02.578662. doi: 10.1101/2024.02.02.578662. bioRxiv. 2024. Update in: Genome Res. 2025 Jul 1;35(7):1621-1632. doi: 10.1101/gr.279380.124. PMID: 38352580 Free PMC article. Updated. Preprint.
Similar articles
-
Spatial domain detection using contrastive self-supervised learning for spatial multi-omics technologies.bioRxiv [Preprint]. 2024 Feb 4:2024.02.02.578662. doi: 10.1101/2024.02.02.578662. bioRxiv. 2024. Update in: Genome Res. 2025 Jul 1;35(7):1621-1632. doi: 10.1101/gr.279380.124. PMID: 38352580 Free PMC article. Updated. Preprint.
-
spaLLM: enhancing spatial domain analysis in multi-omics data through large language model integration.Brief Bioinform. 2025 Jul 2;26(4):bbaf304. doi: 10.1093/bib/bbaf304. Brief Bioinform. 2025. PMID: 40608008 Free PMC article.
-
Unveiling spatial domains from spatial multi-omics data using dual-graph regularized ensemble learning.Commun Biol. 2025 Jun 20;8(1):945. doi: 10.1038/s42003-025-08372-6. Commun Biol. 2025. PMID: 40542177 Free PMC article.
-
Deep learning applications in single-cell genomics and transcriptomics data analysis.Biomed Pharmacother. 2023 Sep;165:115077. doi: 10.1016/j.biopha.2023.115077. Epub 2023 Jul 1. Biomed Pharmacother. 2023. PMID: 37393865
-
Multi-omics approaches: transforming the landscape of natural product isolation.Funct Integr Genomics. 2025 Jun 19;25(1):132. doi: 10.1007/s10142-025-01645-7. Funct Integr Genomics. 2025. PMID: 40537580 Review.
Cited by
-
STimage-1K4M: A histopathology image-gene expression dataset for spatial transcriptomics.ArXiv [Preprint]. 2024 Jun 20:arXiv:2406.06393v2. ArXiv. 2024. PMID: 38947920 Free PMC article. Preprint.
References
-
- Bradski G. 2000. The OpenCV Library. Dr Dobb's Journal of Software Tools 25: 120–123.
-
- Denisenko E, De Kock L, Tan A, Beasley AB, Beilin M, Jones ME, Hou R, Muirí DÓ, Bilic S, Mohan GRKA, et al. 2024. Spatial transcriptomics reveals discrete tumour microenvironments and autocrine loops within ovarian cancer subclones. Nat Commun 15: 2860. 10.1038/s41467-024-47271-y - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
