Epigenome-wide DNA methylation association study of CHIP provides insight into perturbed gene regulation
- PMID: 40393957
- PMCID: PMC12092741
- DOI: 10.1038/s41467-025-59333-w
Epigenome-wide DNA methylation association study of CHIP provides insight into perturbed gene regulation
Abstract
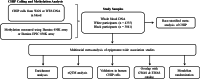
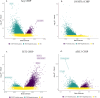
With age, hematopoietic stem cells can acquire somatic mutations in leukemogenic genes that confer a proliferative advantage in a phenomenon termed CHIP. How these mutations result in increased risk for numerous age-related diseases remains poorly understood. We conduct a multiracial meta-analysis of EWAS of CHIP in the Framingham Heart Study, Jackson Heart Study, Cardiovascular Health Study, and Atherosclerosis Risk in Communities cohorts (N = 8196) to elucidate the molecular mechanisms underlying CHIP and illuminate how these changes influence cardiovascular disease risk. We functionally validate the EWAS findings using human hematopoietic stem cell models of CHIP. We then use expression quantitative trait methylation analysis to identify transcriptomic changes associated with CHIP-associated CpGs. Causal inference analyses reveal 261 CHIP-associated CpGs associated with cardiovascular traits and all-cause mortality (FDR adjusted p-value < 0.05). Taken together, our study reports the epigenetic changes impacted by CHIP and their associations with age-related disease outcomes.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: F.P. is an employee of Biomodal. L.M.R serves as a consultant for the NHLBI TOPMed Administrative Coordinating Center (through Westat). P.N. reports research grants from Allelica, Amgen, Apple, Boston Scientific, Genentech / Roche, and Novartis, personal fees from Allelica, Apple, AstraZeneca, Blackstone Life Sciences, Creative Education Concepts, CRISPR Therapeutics, Eli Lilly & Co, Foresite Labs, Genentech / Roche, GV, HeartFlow, Magnet Biomedicine, Merck, and Novartis, scientific advisory board membership of Esperion Therapeutics, Preciseli, TenSixteen Bio, and Tourmaline Bio, scientific co-founder of TenSixteen Bio, equity in MyOme, Preciseli, and TenSixteen Bio, and spousal employment at Vertex Pharmaceuticals, all unrelated to the present work. B.P. serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. The remaining authors – S.K., T.H., J.A., R.J., M.U., N.N., B.Y., J.A.B., M.F., J.B., N.S., D.O., J.F., C.B., L.R., K.C., J.W., A.C., L.L., K.F., N.H., J.M., A.B., and D.L. – declare no competing interests.
Figures

Update of
-
Epigenome-wide DNA Methylation Association Study of CHIP Provides Insight into Perturbed Gene Regulation.Res Sq [Preprint]. 2024 Jul 16:rs.3.rs-4656898. doi: 10.21203/rs.3.rs-4656898/v1. Res Sq. 2024. Update in: Nat Commun. 2025 May 20;16(1):4678. doi: 10.1038/s41467-025-59333-w. PMID: 39070619 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Grants and funding
- HHSN268201100037C/HL/NHLBI NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- U01 HL130114/HL/NHLBI NIH HHS/United States
- U01 HL120393/HL/NHLBI NIH HHS/United States
- R01 HL087652/HL/NHLBI NIH HHS/United States
- HHSN268201800014C/HL/NHLBI NIH HHS/United States
- 75N92022D00004/HL/NHLBI NIH HHS/United States
- N01 HC055222/HL/NHLBI NIH HHS/United States
- HHSN268201500001I/HL/NHLBI NIH HHS/United States
- N01 HC085079/HL/NHLBI NIH HHS/United States
- HHSN268201800011C/HL/NHLBI NIH HHS/United States
- 75N92022D00003/HL/NHLBI NIH HHS/United States
- HHSN268201800015I/HB/NHLBI NIH HHS/United States
- HHSN268201800010I/HB/NHLBI NIH HHS/United States
- HHSN268201800011I/HB/NHLBI NIH HHS/United States
- R01 HL103612/HL/NHLBI NIH HHS/United States
- N01 HC085080/HL/NHLBI NIH HHS/United States
- HHSN268201800012I/HB/NHLBI NIH HHS/United States
- HHSN268201800012C/HL/NHLBI NIH HHS/United States
- T32 GM080178/GM/NIGMS NIH HHS/United States
- 75N92022D00002/HL/NHLBI NIH HHS/United States
- U01 HL080295/HL/NHLBI NIH HHS/United States
- HHSN268201500001C/HL/NHLBI NIH HHS/United States
- N01 HC085082/HL/NHLBI NIH HHS/United States
- HHSN268201800014I/HB/NHLBI NIH HHS/United States
- HHSN268200800007C/HL/NHLBI NIH HHS/United States
- N01 HC085086/HL/NHLBI NIH HHS/United States
- N01 HC085083/HL/NHLBI NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- R01 HL168894/HL/NHLBI NIH HHS/United States
- HHSN268201200036C/HL/NHLBI NIH HHS/United States
- HHSN268201800001C/HL/NHLBI NIH HHS/United States
- N01 HC025195/HL/NHLBI NIH HHS/United States
- HHSN268201800013I/MD/NIMHD NIH HHS/United States
- 75N92021D00006/HL/NHLBI NIH HHS/United States
- 75N92022D00005/HL/NHLBI NIH HHS/United States
- R01 HL117626/HL/NHLBI NIH HHS/United States
- R01 AG023629/AG/NIA NIH HHS/United States
- 75N92022D00001/HL/NHLBI NIH HHS/United States
- R01 HL148050/HL/NHLBI NIH HHS/United States
- N01 HC085081/HL/NHLBI NIH HHS/United States
- U54 DK106829/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources

