Caspase-11 drives macrophage hyperinflammation in models of Polg-related mitochondrial disease
- PMID: 40393978
- PMCID: PMC12092707
- DOI: 10.1038/s41467-025-59907-8
Caspase-11 drives macrophage hyperinflammation in models of Polg-related mitochondrial disease
Abstract
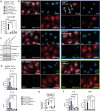
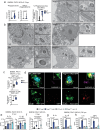
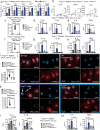
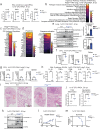
Mitochondrial diseases (MtD) represent a significant public health challenge due to their heterogenous clinical presentation, often severe and progressive symptoms, and lack of effective therapies. Environmental exposures, such bacterial and viral infection, can further compromise mitochondrial function and exacerbate the progression of MtD. However, the underlying immune alterations that enhance immunopathology in MtD remain unclear. Here we employ in vitro and in vivo approaches to clarify the molecular and cellular basis for innate immune hyperactivity in models of polymerase gamma (Polg)-related MtD. We reveal that type I interferon (IFN-I)-mediated upregulation of caspase-11 and guanylate-binding proteins (GBP) increase macrophage sensing of the opportunistic microbe Pseudomonas aeruginosa (PA) in Polg mutant mice. Furthermore, we show that excessive cytokine secretion and activation of pyroptotic cell death pathways contribute to lung inflammation and morbidity after infection with PA. Our work provides a mechanistic framework for understanding innate immune dysregulation in MtD and reveals potential targets for limiting infection- and inflammation-related complications in Polg-related MtD.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



Update of
-
Caspase-11 drives macrophage hyperinflammation in models of Polg-related mitochondrial disease.bioRxiv [Preprint]. 2025 Jan 17:2024.05.11.593693. doi: 10.1101/2024.05.11.593693. bioRxiv. 2025. Update in: Nat Commun. 2025 May 20;16(1):4640. doi: 10.1038/s41467-025-59907-8. PMID: 38798587 Free PMC article. Updated. Preprint.
Similar articles
-
Caspase-11 drives macrophage hyperinflammation in models of Polg-related mitochondrial disease.bioRxiv [Preprint]. 2025 Jan 17:2024.05.11.593693. doi: 10.1101/2024.05.11.593693. bioRxiv. 2025. Update in: Nat Commun. 2025 May 20;16(1):4640. doi: 10.1038/s41467-025-59907-8. PMID: 38798587 Free PMC article. Updated. Preprint.
-
Necrosis drives susceptibility to Mycobacterium tuberculosis in PolgD257A mutator mice.Infect Immun. 2025 Mar 11;93(3):e0032424. doi: 10.1128/iai.00324-24. Epub 2025 Feb 19. Infect Immun. 2025. PMID: 39969190 Free PMC article.
-
The importance of Fcγ and C-type lectin receptors in host immune responses during Pneumocystis pneumonia.Infect Immun. 2025 Feb 18;93(2):e0027624. doi: 10.1128/iai.00276-24. Epub 2024 Dec 31. Infect Immun. 2025. PMID: 39745390 Free PMC article.
-
Epilepsy due to mutations in the mitochondrial polymerase gamma (POLG) gene: A clinical and molecular genetic review.Epilepsia. 2016 Oct;57(10):1531-1545. doi: 10.1111/epi.13508. Epub 2016 Aug 24. Epilepsia. 2016. PMID: 27554452
-
Antibiotic strategies for eradicating Pseudomonas aeruginosa in people with cystic fibrosis.Cochrane Database Syst Rev. 2017 Apr 25;4(4):CD004197. doi: 10.1002/14651858.CD004197.pub5. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2023 Jun 2;6:CD004197. doi: 10.1002/14651858.CD004197.pub6. PMID: 28440853 Free PMC article. Updated.
Cited by
-
Necrosis drives susceptibility to Mycobacterium tuberculosis in PolgD257A mutator mice.bioRxiv [Preprint]. 2024 Dec 2:2024.07.17.603991. doi: 10.1101/2024.07.17.603991. bioRxiv. 2024. Update in: Infect Immun. 2025 Mar 11;93(3):e0032424. doi: 10.1128/iai.00324-24. PMID: 39091776 Free PMC article. Updated. Preprint.
-
Tipping the balance: innate and adaptive immunity in mitochondrial disease.Curr Opin Immunol. 2025 Aug;95:102566. doi: 10.1016/j.coi.2025.102566. Epub 2025 May 26. Curr Opin Immunol. 2025. PMID: 40424975 Review.
References
-
- Gorman, G. S. et al. Mitochondrial diseases. Nat. Rev. Dis. Prim.2, 16080 (2016). - PubMed
-
- Suomalainen, A. & Battersby, B. J. Mitochondrial diseases: the contribution of organelle stress responses to pathology. Nat. Rev. Mol. Cell Biol.19, 77–92 (2018). - PubMed
-
- Russell, O. M., Gorman, G. S., Lightowlers, R. N. & Turnbull, D. M. Mitochondrial diseases: hope for the future. Cell181, 168–188 (2020). - PubMed
-
- Walker, M. A. et al. Predisposition to infection and SIRS in mitochondrial disorders: 8 years’ experience in an academic center. J. Allergy Clin. Immunol. Pr.2, 465–468, 468.e1 (2014). - PubMed
MeSH terms
Substances
Grants and funding
- W81XWH-17-1-0052/United States Department of Defense | United States Army | Army Medical Command | Congressionally Directed Medical Research Programs (CDMRP)
- P30 CA034196/CA/NCI NIH HHS/United States
- R01HL148153/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- F31AI179168/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- U54 OD030187/OD/NIH HHS/United States
- F31 AI179168/AI/NIAID NIH HHS/United States
- W81XWH-20-1-0150/United States Department of Defense | United States Army | Army Medical Command | Congressionally Directed Medical Research Programs (CDMRP)
- R01 HL148153/HL/NHLBI NIH HHS/United States
- U54OD030187/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- 825908/American Heart Association (American Heart Association, Inc.)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

