CREBBP inactivation sensitizes B cell acute lymphoblastic leukemia to ferroptotic cell death upon BCL2 inhibition
- PMID: 40393984
- PMCID: PMC12092839
- DOI: 10.1038/s41467-025-59531-6
CREBBP inactivation sensitizes B cell acute lymphoblastic leukemia to ferroptotic cell death upon BCL2 inhibition
Abstract
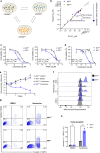
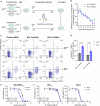
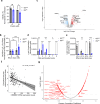
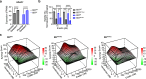
B-cell acute lymphoblastic leukemia (B-ALL) is a leading cause of death in childhood and outcomes in adults remain dismal. There is therefore an urgent clinical need for therapies that target the highest risk cases. Mutations in the histone acetyltransferase CREBBP confer high-risk and increased chemoresistance in ALL. Performing a targeted drug-screen in isogenic human cell lines, we identify a number of small molecules that specifically target CREBBP-mutated B-ALL, the most potent being the BCL2-inhibitor Venetoclax. Of note, this acts through a non-canonical mechanism resulting in ferroptotic rather than apoptotic cell death. CREBBP-mutated cell lines show differences in cell-cycle, metabolism, lipid composition and response to oxidative stress, predisposing them to ferroptosis, which are further dysregulated upon acquisition of Venetoclax resistance. Lastly, small-molecule inhibition of CREBBP pharmacocopies CREBBP-mutation, sensitizing B-ALL cells, regardless of genotype, to Venetoclax-induced ferroptosis in-vitro and in-vivo, providing a promising drug combination for broader clinical translation in B-ALL.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: N.N. is a former employee of the Walter and Eliza Hall Institute which receives milestone and royalty payments related to Venetoclax. N.N. received payments from WEHI related to Venetoclax. The combination of BCL2 and CREBBP/EP300 inhibitors in ALL is protected under Cambridge Enterprise patent PCT/GB2024/052398 (S.E.R. and B.P.J.H. listed inventors). The remaining authors declare no competing interests.
Figures




References
-
- Hunger, S. P. & Mullighan, C. G. Acute lymphoblastic leukemia in children. N. Engl. J. Med.373, 1541–1552 (2015). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

