APOBEC affects tumor evolution and age at onset of lung cancer in smokers
- PMID: 40394004
- PMCID: PMC12092836
- DOI: 10.1038/s41467-025-59923-8
APOBEC affects tumor evolution and age at onset of lung cancer in smokers
Abstract
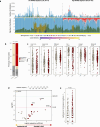
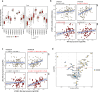
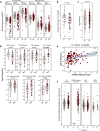
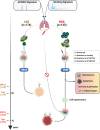
Most solid tumors harbor somatic mutations attributed to off-target activities of APOBEC3A (A3A) and/or APOBEC3B (A3B). However, how APOBEC3A/B enzymes affect tumor evolution in the presence of exogenous mutagenic processes is largely unknown. Here, multi-omics profiling of 309 lung cancers from smokers identifies two subtypes defined by low (LAS) and high (HAS) APOBEC mutagenesis. LAS are enriched for A3B-like mutagenesis and KRAS mutations; HAS for A3A-like mutagenesis and TP53 mutations. Compared to LAS, HAS have older age at onset and high proportions of newly generated progenitor-like cells likely due to the combined tobacco smoking- and APOBEC3A-associated DNA damage and apoptosis. Consistently, HAS exhibit high expression of pulmonary healing signaling pathway, stemness markers, distal cell-of-origin, more neoantigens, slower clonal expansion, but no smoking-associated genomic/epigenomic changes. With validation in 184 lung tumor samples, these findings show how heterogeneity in mutational burden across co-occurring mutational processes and cell types contributes to tumor development.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: L.B.A. is a compensated consultant and has equity interest in io9, LLC. His spouse is an employee of Biotheranostics, Inc. L.B.A. is also an inventor of a US Patent 10,776,718 for source identification by non-negative matrix factorization. E.N.B. and L.B.A. declare U.S. provisional patent applications with serial numbers 63/289,601 and 63/269,033. L.B.A. also declares U.S. provisional patent applications with serial numbers: 63/366,392; 63/367,846; and 63/412,835. All other authors declare no competing interests.
Figures

Update of
-
APOBEC shapes tumor evolution and age at onset of lung cancer in smokers.bioRxiv [Preprint]. 2024 Apr 3:2024.04.02.587805. doi: 10.1101/2024.04.02.587805. bioRxiv. 2024. Update in: Nat Commun. 2025 May 21;16(1):4711. doi: 10.1038/s41467-025-59923-8. PMID: 38617360 Free PMC article. Updated. Preprint.
Similar articles
-
APOBEC shapes tumor evolution and age at onset of lung cancer in smokers.bioRxiv [Preprint]. 2024 Apr 3:2024.04.02.587805. doi: 10.1101/2024.04.02.587805. bioRxiv. 2024. Update in: Nat Commun. 2025 May 21;16(1):4711. doi: 10.1038/s41467-025-59923-8. PMID: 38617360 Free PMC article. Updated. Preprint.
-
APOBEC3A is a prominent cytidine deaminase in breast cancer.PLoS Genet. 2019 Dec 16;15(12):e1008545. doi: 10.1371/journal.pgen.1008545. eCollection 2019 Dec. PLoS Genet. 2019. PMID: 31841499 Free PMC article.
-
APOBEC3 Activity Promotes the Survival and Evolution of Drug-Tolerant Persister Cells during EGFR Inhibitor Resistance in Lung Cancer.Cancer Res Commun. 2025 May 1;5(5):825-840. doi: 10.1158/2767-9764.CRC-24-0442. Cancer Res Commun. 2025. PMID: 40323013 Free PMC article.
-
Regulation, functional impact, and therapeutic targeting of APOBEC3A in cancer.DNA Repair (Amst). 2024 Sep;141:103734. doi: 10.1016/j.dnarep.2024.103734. Epub 2024 Jul 20. DNA Repair (Amst). 2024. PMID: 39047499 Review.
-
Roles of APOBEC3A and APOBEC3B in Human Papillomavirus Infection and Disease Progression.Viruses. 2017 Aug 21;9(8):233. doi: 10.3390/v9080233. Viruses. 2017. PMID: 28825669 Free PMC article. Review.
Cited by
-
Tobacco smoke carcinogens exacerbate APOBEC mutagenesis and carcinogenesis.Res Sq [Preprint]. 2025 Jun 3:rs.3.rs-5843684. doi: 10.21203/rs.3.rs-5843684/v1. Res Sq. 2025. PMID: 40502742 Free PMC article. Preprint.
-
Tobacco smoke carcinogens exacerbate APOBEC mutagenesis and carcinogenesis.bioRxiv [Preprint]. 2025 Jan 22:2025.01.18.633716. doi: 10.1101/2025.01.18.633716. bioRxiv. 2025. PMID: 39896515 Free PMC article. Preprint.
-
The mutagenic forces shaping the genomic landscape of lung cancer in never smokers.medRxiv [Preprint]. 2024 May 17:2024.05.15.24307318. doi: 10.1101/2024.05.15.24307318. medRxiv. 2024. PMID: 38798417 Free PMC article. Preprint.
-
The mutagenic forces shaping the genomes of lung cancer in never smokers.Nature. 2025 Aug;644(8075):133-144. doi: 10.1038/s41586-025-09219-0. Epub 2025 Jul 2. Nature. 2025. PMID: 40604281
References
-
- Koh, G., Degasperi, A., Zou, X., Momen, S. & Nik-Zainal, S. Mutational signatures: emerging concepts, caveats and clinical applications. Nat. Rev. Cancer21, 619–637 (2021). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

