Histone methyltransferase ASH1L primes metastases and metabolic reprogramming of macrophages in the bone niche
- PMID: 40394007
- PMCID: PMC12092585
- DOI: 10.1038/s41467-025-59381-2
Histone methyltransferase ASH1L primes metastases and metabolic reprogramming of macrophages in the bone niche
Abstract
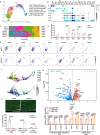
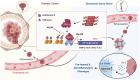
Bone metastasis is a major cause of cancer death; however, the epigenetic determinants driving this process remain elusive. Here, we report that histone methyltransferase ASH1L is genetically amplified and is required for bone metastasis in men with prostate cancer. ASH1L rewires histone methylations and cooperates with HIF-1α to induce pro-metastatic transcriptome in invading cancer cells, resulting in monocyte differentiation into lipid-associated macrophage (LA-TAM) and enhancing their pro-tumoral phenotype in the metastatic bone niche. We identified IGF-2 as a direct target of ASH1L/HIF-1α and mediates LA-TAMs' differentiation and phenotypic changes by reprogramming oxidative phosphorylation. Pharmacologic inhibition of the ASH1L-HIF-1α-macrophages axis elicits robust anti-metastasis responses in preclinical models. Our study demonstrates epigenetic alterations in cancer cells reprogram metabolism and features of myeloid components, facilitating metastatic outgrowth. It establishes ASH1L as an epigenetic driver priming metastasis and macrophage plasticity in the bone niche, providing a bona fide therapeutic target in metastatic malignancies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
-
- Gupta, G. P. & Massague, J. Cancer metastasis: building a framework. Cell127, 679–695 (2006). - PubMed
MeSH terms
Substances
Grants and funding
- FP00016492/Prostate Cancer Foundation (PCF)
- R01 CA269646/CA/NCI NIH HHS/United States
- R01 CA181196/CA/NCI NIH HHS/United States
- R01 CA278889/CA/NCI NIH HHS/United States
- PC230358/U.S. Department of Defense (United States Department of Defense)
- CA166051/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- U54 CA274220/CA/NCI NIH HHS/United States
- R01 CA166051/CA/NCI NIH HHS/United States
- S10 OD024977/OD/NIH HHS/United States
- RR190021/Cancer Prevention and Research Institute of Texas (Cancer Prevention Research Institute of Texas)
- CA269140/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01 CA244144/CA/NCI NIH HHS/United States
- R01 CA247992/CA/NCI NIH HHS/United States
- R01 CA269140/CA/NCI NIH HHS/United States
- R01 CA275990/CA/NCI NIH HHS/United States
- CA278889/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- CA275990/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

