Sphingolipid metabolism-related genes as diagnostic markers in pneumonia-induced sepsis: the AUG model
- PMID: 40394027
- PMCID: PMC12092762
- DOI: 10.1038/s41598-025-01150-8
Sphingolipid metabolism-related genes as diagnostic markers in pneumonia-induced sepsis: the AUG model
Abstract
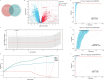
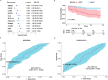
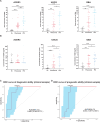
Pneumonia-induced sepsis (PIS) is a life-threatening condition with high mortality rates, necessitating the identification of biomarkers and therapeutic targets. Sphingolipid, particularly ceramides, are pivotal in modulating immune responses and determining cell fate. In this study, we identified a novel gene signature related to sphingolipid metabolism, comprising ACER3, UGCG, and GBA, which are key enzymes involved in the synthesis and metabolism of ceramides. This signature, termed the "AUG model", demonstrated strong diagnostic performance and modest prognostic efficacy across both training (GSE65682) and validation (E-MTAB-1548 and E-MTAB-5273) datasets. A clinical cohort comprising 20 PIS patients, 31 pneumonia cases, and 11 healthy controls further validated the increased expression of AUG genes at both mRNA and protein levels in peripheral blood samples upon admission. Our comprehensive analysis of bulk and single-cell transcriptome datasets revealed that these genes are implicated in immune cell death pathways, including autophagy and apoptosis. Additionally, cell-communication analysis indicated that enhanced macrophage migration inhibitory factor (MIF) signaling may be associated with dysregulated sphingolipid metabolism, potentially driving the inflammatory cascade. This study identifies a novel predictive model for PIS, highlighting the role of sphingolipid metabolism-related genes in disease progression and suggesting potential therapeutic targets for sepsis management.
Keywords: MIF signaling; Pneumonia-induced sepsis; Programmed cell death; Sphingolipid metabolism.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethics approval and consent to participate: This study received ethical approval from the ethics committee of the School of Medicine, Xiamen University (Approval No. XDYX202302K06). Before sample collection, consent was obtained from the guardians. Consent for publication: The authors have consent for publication.
Figures



Similar articles
-
Integrating Single-Cell and Bulk RNA Sequencing Data to Explore Sphingolipid Metabolism Molecular Signatures in Ovarian Cancer Prognosis: an Original Study.Int J Med Sci. 2025 Mar 24;22(8):1958-1977. doi: 10.7150/ijms.107391. eCollection 2025. Int J Med Sci. 2025. PMID: 40225866 Free PMC article.
-
Identification of Hub Genes and Key Pathways Associated with Sepsis Progression Using Weighted Gene Co-Expression Network Analysis and Machine Learning.Int J Mol Sci. 2025 May 7;26(9):4433. doi: 10.3390/ijms26094433. Int J Mol Sci. 2025. PMID: 40362669 Free PMC article.
-
Patterns of gene expression in peripheral blood mononuclear cells and outcomes from patients with sepsis secondary to community acquired pneumonia.PLoS One. 2014 Mar 25;9(3):e91886. doi: 10.1371/journal.pone.0091886. eCollection 2014. PLoS One. 2014. PMID: 24667684 Free PMC article.
-
The key role of sphingolipid metabolism in cancer: New therapeutic targets, diagnostic and prognostic values, and anti-tumor immunotherapy resistance.Front Oncol. 2022 Jul 27;12:941643. doi: 10.3389/fonc.2022.941643. eCollection 2022. Front Oncol. 2022. PMID: 35965565 Free PMC article. Review.
-
Aquaporins in sepsis- an update.Front Immunol. 2024 Oct 31;15:1495206. doi: 10.3389/fimmu.2024.1495206. eCollection 2024. Front Immunol. 2024. PMID: 39544938 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

