SKSR1 identified as key virulence factor in Cryptosporidium by genetic crossing
- PMID: 40394032
- PMCID: PMC12092579
- DOI: 10.1038/s41467-025-60088-7
SKSR1 identified as key virulence factor in Cryptosporidium by genetic crossing
Abstract
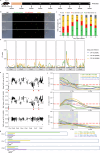
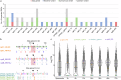
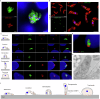
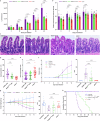
Cryptosporidium is a major cause of severe diarrhea. Although Cryptosporidium isolates exhibit significant differences in infectivity and virulence, the genetic determinants for these traits are not clear. In this study, we use classical genetics to cross two Cryptosporidium parvum isolates of different virulence and use bulk segregant analysis of whole-genome sequences from the progeny to identify quantitative trait loci (QTL) associated with Cryptosporidium infectivity and virulence. Of the 23 genes in three QTL, two have loss-of-function mutations in the low-virulence isolates, including the SKSR1 gene encoding a variant secretory protein. Deletion of the SKSR1 gene or expression of the frame-shifted sequence reduces the pathogenicity of the virulent isolate. SKSR1 is expressed in small granules and secreted into the parasite-host interface during invasion. These results demonstrate that SKSR1 is an important virulence factor in Cryptosporidium, and suggest that the extended SKSR protein family, encoded by clusters of subtelomeric genes, may contribute to pathogenesis.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Involvement of a variant secretory protein in virulence of emerging Cryptosporidium parvum subtypes.Virulence. 2025 Dec;16(1):2514077. doi: 10.1080/21505594.2025.2514077. Epub 2025 Jun 4. Virulence. 2025. PMID: 40465693 Free PMC article.
-
Genetic basis for virulence differences of various Cryptosporidium parvum carcinogenic isolates.Sci Rep. 2020 Apr 30;10(1):7316. doi: 10.1038/s41598-020-64370-0. Sci Rep. 2020. PMID: 32355272 Free PMC article.
-
Cryptosporidium virulence determinants--are we there yet?Int J Parasitol. 2002 May;32(5):517-25. doi: 10.1016/s0020-7519(01)00356-3. Int J Parasitol. 2002. PMID: 11943224 Review.
-
Comparative genomic analysis reveals occurrence of genetic recombination in virulent Cryptosporidium hominis subtypes and telomeric gene duplications in Cryptosporidium parvum.BMC Genomics. 2015 Apr 18;16(1):320. doi: 10.1186/s12864-015-1517-1. BMC Genomics. 2015. PMID: 25903370 Free PMC article.
-
Cryptosporidium pathogenicity and virulence.Clin Microbiol Rev. 2013 Jan;26(1):115-34. doi: 10.1128/CMR.00076-12. Clin Microbiol Rev. 2013. PMID: 23297262 Free PMC article. Review.
References
-
- Kotloff, K. L. et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet382, 209–222 (2013). - PubMed
-
- Feng, Y., Ryan, U. M. & Xiao, L. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol.34, 997–1011 (2018). - PubMed
MeSH terms
Substances
Grants and funding
- 32030109/National Natural Science Foundation of China (National Science Foundation of China)
- 32150710530/National Natural Science Foundation of China (National Science Foundation of China)
- 31972697/National Natural Science Foundation of China (National Science Foundation of China)
- 31820103014/National Natural Science Foundation of China (National Science Foundation of China)
- 31972697/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Medical

