Targeting epigenetic regulators to overcome drug resistance in the emerging human fungal pathogen Candida auris
- PMID: 40394068
- PMCID: PMC12092656
- DOI: 10.1038/s41467-025-59898-6
Targeting epigenetic regulators to overcome drug resistance in the emerging human fungal pathogen Candida auris
Abstract
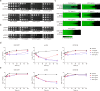
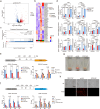
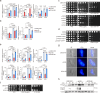
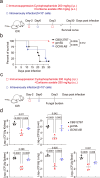
The rise of drug-resistant fungal species, such as Candida auris, poses a serious threat to global health, with mortality rates exceeding 40% and resistance rates surpassing 90%. The limited arsenal of effective antifungal agents underscores the urgent need for novel strategies. Here, we systematically evaluate the role of histone H3 post-translational modifications in C. auris drug resistance, focusing on acetylation mediated by Gcn5 and Rtt109, and methylation mediated by Set1, Set2, and Dot1. Mutants deficient in these enzymes exhibit varying degrees of antifungal drug sensitivity. Notably, we discover that GCN5 depletion and the subsequent loss of histone H3 acetylation downregulates key genes involved in ergosterol biosynthesis and drug efflux, resulting in increased susceptibility to azoles and polyenes. Additionally, Gcn5 regulates cell wall integrity and echinocandin resistance through the calcineurin signaling pathway and transcription factor Cas5. In infection models using Galleria mellonella and immunocompromised mice, GCN5 deletion significantly reduces the virulence of C. auris. Furthermore, the Gcn5 inhibitor CPTH2 synergizes with caspofungin in vitro and in vivo without notable toxicity. These findings highlight the critical role of Gcn5 in the resistance and pathogenicity of C. auris, positioning it as a promising therapeutic target for combating invasive fungal infections.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Jermy, A. Stop neglecting fungi. Nat. Microbiol.2, 17120 (2017). - PubMed
-
- Logan, C., Martin-Loeches, I. & Bicanic, T. Invasive candidiasis in critical care: challenges and future directions. Intensive Care Med.46, 2001–2014 (2020). - PubMed
-
- Kenters, N. et al. Control of Candida auris in healthcare institutions: outcome of an international society for antimicrobial chemotherapy expert meeting. Int. J. Antimicrob. Agents54, 400–406 (2019). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

