In depth transcriptomic profiling defines a landscape of dysfunctional immune responses in patients with VEXAS syndrome
- PMID: 40394087
- PMCID: PMC12092610
- DOI: 10.1038/s41467-025-59890-0
In depth transcriptomic profiling defines a landscape of dysfunctional immune responses in patients with VEXAS syndrome
Abstract
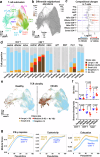
VEXAS (Vacuoles, E1 enzyme, X-linked, Autoinflammatory, Somatic) syndrome is caused by inactivating somatic mutations in the UBA1 gene. Here, we characterize the immunological landscape of VEXAS syndrome by performing multi-omics single-cell RNA analysis, cytokine multiplex assays, and in vitro functional assays on patients' peripheral blood. Our data reveals a broad immune system activation with upregulation of multiple inflammatory response pathways and proinflammatory cytokines. Unexpectedly, we find that monocytes have dysfunctional features irrespective of UBA1 mutation status, exhibiting impaired efferocytosis and blunted cytokine production in vitro. In contrast, UBA1-mutated NK cells show an upregulation of the inflammation pathways and enhanced cytotoxicity. Within the lymphocyte subsets, predominantly UBA1 wild-type, we identify clonal expansion of effector memory CD8+ T cells and skewed B cell differentiation with loss of transitional B cells and expansion of plasmablasts. Thus, our analysis indicates that VEXAS syndrome is characterized by profound alterations in both adaptive and innate immune systems, accounting for the complex pathophysiology of the disease, and provides a basis to understand the marked clinical heterogeneity and variable disease course.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

