A novel luciferase-based assay for quantifying coronavirus-induced syncytia
- PMID: 40394090
- PMCID: PMC12092680
- DOI: 10.1038/s41598-025-02037-4
A novel luciferase-based assay for quantifying coronavirus-induced syncytia
Abstract
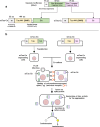
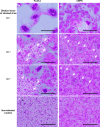
Coronaviruses can induce cell‒cell fusion that results in the formation of multinucleated syncytia through the interaction of viral spike proteins with host cell receptors. Quantifying syncytial formation is crucial for screening potential efficacious antiviral compounds. However, some traditional methods for syncytial quantification are often labor-intensive and limited by a low-throughput capacity. Therefore, we developed a novel high-throughput assay for the efficient quantification of syncytial formation induced by feline coronavirus (FCoV) and SARS-CoV-2. This assay, which is based on the split luciferase system, utilizes a split Gaussia luciferase (Gluc) system. In this system, fragments of Gluc are fused to the multimerizing Tau protein to reconstitute enzymatic activity upon cell fusion. In this study, the activity of the reconstituted luciferase was measured in 20 µL of culture medium to efficiently quantify syncytial formation induced by FCoV and SARS-CoV-2. Our findings demonstrate that this assay can accelerate the discovery of antiviral drugs that target coronaviruses.
Keywords: Coronaviruses; Drug discovery; Drug screening; SARS-CoV-2; Syncytium quantification; Tau proteins.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures





References
-
- Gruters, R. A. et al. Interference with HIV-induced syncytium formation and viral infectivity by inhibitors of trimming glucosidase. Nature330, 74–77. 10.1038/330074a0 (1987). - PubMed
-
- Taguchi, F., Ikeda, T. & Shida, H. Molecular cloning and expression of a Spike protein of neurovirulent murine coronavirus JHMV variant cl-2. J. Gen. Virol.73 (Pt 5), 1065–1072. 10.1099/0022-1317-73-5-1065 (1992). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

