D-optimal design model and biosynthetic pathway for gentamicin production by Micromonospora purpureochromogenes NRRL B-16094
- PMID: 40394500
- PMCID: PMC12090562
- DOI: 10.1186/s12866-025-04001-8
D-optimal design model and biosynthetic pathway for gentamicin production by Micromonospora purpureochromogenes NRRL B-16094
Abstract
Background: Micromonospora purpureochromogenes NRRL B-16094, a natural producer of gentamicin (GEN), a 5,6-diglycosylated 2-dexoystreptamine-aminoglycoside antibiotic (2DOS-AGA) broad-spectrum bactericidal activity. In literature, limited studies are concerned with the biosynthetic route and various cultural conditions influencing GEN production.
Methods: Therefore, this study aimed to explore the GEN biosynthesis pathway and compare it to that of fortimicin and kanamycin. In addition, four key environmental conditions influencing GEN production were statistically optimized using response surface D-optimal design (DOD). Herein, the biosynthetic pathway of GEN was proposed based on the biochemistry of the identified genes/proteins within the gene cluster. Comparing the GEN-biosynthetic gene cluster to that of kanamycin and fortimicin suggested that gentamicin biosynthesis could have originated from a combination of biosynthetic pathways of both antibiotics.
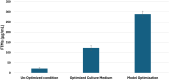
Results: For the optimization experiments, culture media 4 (CM4) and 6 (CM6) gave the highest specific productivity at 6.36 and 3.80 µg/mg, respectively. A DOD quadratic model was successfully generated to optimize four key environmental factors. Predicted and experimentally confirmed optimized factors were an initial pH of 7, an incubation temperature of 30˚C, and an agitation of 300 rpm for 10 days. This resulted in a 13.5-fold increase (289.5 µg/mL) over that produced by the basic CM1 production medium (21.4 µg/mL) and 2.4 times (over that obtained by CM4 (123.7 µg/mL) as verified by HPLC analysis.
Conclusion: DOD is an efficient tool for optimizing GEN. Accordingly, the optimized conditions are highly advisable during the scaling up of GEN production by M. purpureochromogenes NRRL B-16094.
Keywords: D-optimal design Micromonospora purpureochromogenes NRRL B-16094; Fortimicin; Gentamicin; Kanamycin.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: Not applicable. Consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





Similar articles
-
Deciphering hygromycin B biosynthetic pathway and D-optimal design for production optimization.World J Microbiol Biotechnol. 2025 Apr 28;41(5):155. doi: 10.1007/s11274-025-04364-0. World J Microbiol Biotechnol. 2025. PMID: 40289225
-
Response Surface D-Optimal Design for Optimizing Fortimicins Production by Micromonospora olivasterospora and New Synergistic Fortimicin-A-Antibiotic Combinations.Curr Microbiol. 2025 Jan 3;82(2):68. doi: 10.1007/s00284-024-04049-1. Curr Microbiol. 2025. PMID: 39753822
-
Characterization of a key aminoglycoside phosphotransferase in gentamicin biosynthesis.Bioorg Med Chem Lett. 2013 Mar 1;23(5):1438-41. doi: 10.1016/j.bmcl.2012.12.064. Epub 2013 Jan 3. Bioorg Med Chem Lett. 2013. PMID: 23339967
-
Microbial biosynthesis and applications of gentamicin: a critical appraisal.Crit Rev Biotechnol. 2008;28(3):173-212. doi: 10.1080/07388550802262197. Crit Rev Biotechnol. 2008. PMID: 18937107 Review.
-
Biosynthetic genes for aminoglycoside antibiotics.J Antibiot (Tokyo). 2009 Sep;62(9):471-81. doi: 10.1038/ja.2009.76. Epub 2009 Jul 31. J Antibiot (Tokyo). 2009. PMID: 19644520 Review.
References
-
- Piepersberg W, Aboshanab KM, Schmidt-Beißner H, Wehmeier UF. The biochemistry and genetics of aminoglycoside producers. Aminoglycoside antibiotics. New York, USA: Wiley; 2007. pp. 15–118.
-
- Wehmeier UF, Piepersberg W. Enzymology of aminoglycoside biosynthesis-deduction from gene clusters. Methods Enzymol. 2009;459:459–91. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

