Genetic influence of the brain imaging phenotypes, brain and cerebrospinal fluid metabolites and brain genes on migraine subtypes: a Mendelian randomization and multi-omics study
- PMID: 40394501
- PMCID: PMC12090607
- DOI: 10.1186/s10194-025-02063-7
Genetic influence of the brain imaging phenotypes, brain and cerebrospinal fluid metabolites and brain genes on migraine subtypes: a Mendelian randomization and multi-omics study
Abstract
Background: Migraine is a complex neurological disorder with high prevalence but unclear pathogenesis. Numerous studies have suggested that migraine is associated with alterations in brain imaging phenotypes (BIPs) and dysregulation of cerebrospinal fluid (CSF) and brain metabolism; however, causal evidence remains limited. Mendelian randomization (MR) offers a powerful approach for inferring causality using genetic instruments.
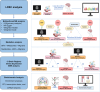
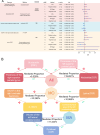
Methods: Firstly, we conducted linkage disequilibrium score regression (LDSC) to evaluate genetic correlations between migraine, including the migraine with aura (MA) and migraine without aura (MO) subtypes, and BIPs, CSF, and brain metabolites. Traits that showed genetic correlations with migraine, MA, or MO were retained for subsequent MR analysis with the corresponding migraine phenotype. Traits showing significant correlations were analyzed using bidirectional two-sample MR (TSMR), followed by two-step TSMR to identify cross-omics mediation effects. Additionally, We also applied summary-data-based MR (SMR) to detect brain-region-specific genes with potential causal effects. Enrichment analyses (KEGG, GO, PPI, transcription factor, and miRNA networks) were conducted to further explore underlying mechanisms.
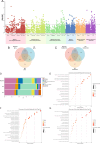
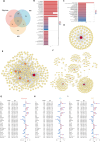
Results: LDSC identified significant genetic correlations with 73 BIPs and 40 metabolites for overall migraine, 71 BIPs and 37 metabolites for MA, and 49 BIPs and 62 metabolites for MO. Enrichment analysis revealed that genetically associated metabolites were predominantly involved in amino acid metabolic pathways. TSMR identified 6 BIPs and 2 metabolites causally linked to overall migraine, 3 BIPs and 3 metabolites to MA, and 2 BIPs and 5 metabolites to MO. Most migraine-related BIPs mapped to the parietal lobe. Reverse MR analysis showed that overall migraine causally influenced 4 BIPs and 3 metabolites, while MA and MO affected 1 BIP and 1 metabolite, and 3 BIPs and 1 metabolite, respectively. Mediation analysis revealed five significant mediation pathways were identified. SMR analysis identified FAM83B and CIB2 consistently showing inhibitory effects across most regions. Enrichment analysis showed that these genes were predominantly involved in immune activation and cell adhesion.
Conclusions: Our study integrates cross-omics analyses to investigate the causal links between brain structure, metabolic alterations, gene expression, and migraine including its MA and MO subtypes. These findings provide novel insights into the pathophysiological mechanisms and potential targets for intervention across migraine subtypes.
Keywords: Brain and cerebrospinal fluid metabolites; Brain imaging phenotypes; Genome-wide association; Mendelian randomization; Migraine; Multi-omics.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: The data for this investigation were acquired from previously published studies and public sources, negating the need for further ethical approval. Competing interests: The authors declare no competing interests.
Figures

References
-
- Headache Classification Committee of the International Headache Society (IHS) The International Classification of Headache Disorders, 3rd edition. Cephalalgia (2018) 38:1-211 - PubMed
-
- Steiner TJ, Stovner LJ (2023) Global epidemiology of migraine and its implications for public health and health policy. Nat Rev Neurol 19:109–117 - PubMed
-
- Dodick DW (2018) Migraine Lancet 391:1315–1330 - PubMed
-
- Lai KL, Niddam DM (2020) Brain metabolism and structure in chronic migraine. Curr Pain Headache Rep 24:69 - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

