Optimized expression of oxazolomycins in engineered Streptomyces longshengensis and their activity evaluation
- PMID: 40394577
- PMCID: PMC12090451
- DOI: 10.1186/s12934-025-02726-9
Optimized expression of oxazolomycins in engineered Streptomyces longshengensis and their activity evaluation
Abstract
Background: To cope with the growing number of severe diseases and intractable pathogens, drug innovation in both chemical structures and pharmacological efficiency has become an imperative global mission. Oxazolomycins are a unique family of polyketide-polypeptide antibiotics from Streptomyces with diverse functional groups in their structures, conferring them multifarious activities. But further development into clinical applications has been hindered for decades for many reasons. Among them, the yield improvement is a critical basis for activity evaluation and drug-like property optimization. This study aims to enhance the production of oxazolomycins in Streptomyces longshengensis through metabolic engineering and evaluate their bioactivity against clinically relevant pathogens.
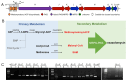
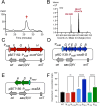
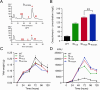
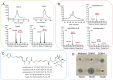
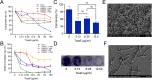
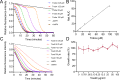
Results: Co-transcriptional analyses suggested that two operons (the transcriptional unit from gene oxaG to oxaB, and that from gene oxaH to oxaQ) could be included in the oxazolomycin biosynthetic gene cluster (oxa BGC) of S. longshengensis. So a strategy was designed to replace the native promoter regions between oxaG and oxaH with constitutive promoters Pneo and PkasO* following functional module evaluation. In the resultant strain (SLOE), the production of oxazolomycin component Toxa5 was increased to 4-fold of that in the wild-type strain. Accordingly, the transcription of all related genes in oxa was clearly promoted. SLOE was then subjected to sublethal dose of gentamicin to induce mutagenesis for optimizing the genetic background, generating a resistant mutant SLROE. With the introduction of transporter genes (ozmS and oxaA) into SLROE, 175 mg/L of Toxa5 was achieved, representing the highest yield in shake-flask fermentation to the best of our knowledge. Finally, the purified Toxa5 showed significant inhibition on the growth of clinically important Gram-negative pathogenic bacterium, Pseudomonas aeruginosa, and the biofilm formation of Bacillus subtilis. Intriguingly, an unprecedented antioxidant activity was also demonstrated.
Conclusions: An oxazolomycin high-producing system of S. longshengensis was established by employing genetic engineering strategies to facilitate the bioactivity exploitation. Oxazolomycin Toxa5 showed interesting inhibitory effects against multiple Gram-negative and -positive pathogens as well as antioxidant capacity, indicating its great potential in clinical applications. The findings provide an efficient strategy for the overproduction and activity evaluation of oxazolomycins.
Keywords: Streptomyces longshengensis; Bioactivity; Biosynthesis; Oxazolomycin.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

References
-
- van Wezel GP, McDowall KJ. The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat Prod Rep. 2011;28(7):1311–33. - PubMed
-
- Li Y, Meng X, Li D, Xia X, Zhang J, Chen Y, Tan H. NeoI represents a group of transcriptional repressors regulating the biosynthesis of multiple aminoglycosides. Sci China Life Sci. 2024;67(12):2761–70. - PubMed
-
- Li Y, Guan H, Li J, Zhang J, Wang Y, Li J, Tan H. An intricate regulation of WblA controlling production of silent Tylosin analogues and abolishment of expressible Nikkomycin. Sci China Life Sci. 2023;66(3):612–25. - PubMed
-
- Zhang J, Tan H. Microbial quorum sensing signaling molecules and their roles in the biosynthesis of natural products. Sci China Life Sci. 2023;66(10):2429–32. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

