The AP2/ERF transcription factor GmTINY mediates ethylene regulation of Rhg1-conferred resistance against soybean cyst nematode
- PMID: 40394901
- PMCID: PMC12281220
- DOI: 10.1016/j.xplc.2025.101378
The AP2/ERF transcription factor GmTINY mediates ethylene regulation of Rhg1-conferred resistance against soybean cyst nematode
Abstract
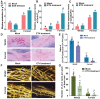
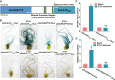
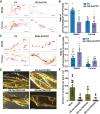
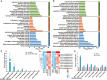
The soybean complex resistance locus Rhg1 is a primary contributor to the control of soybean cyst nematode (Heterodera glycines; SCN), a major threat to soybean production worldwide. Two genes within this locus, GmAATRhg1 and GmSNAP18, are upregulated during SCN infection. In this study, we found that SCN-resistant soybean varieties exhibit elevated ethylene (ETH) production upon infection. Exogenous ETH application further increased GmAATRhg1 and GmSNAP18 transcript levels and enhanced the SCN-resistance response. These genes are divergently transcribed from a shared promoter containing three ERELEE4 ethylene-responsive elements (AATTCAAA). Using yeast one-hybrid screening, we identified the AP2/ERF transcription factor GmTINY (Glyma.01G147600) as an ERELEE4-binding protein. GmTINY is localized to the nucleus, and its expression is induced by both ETH treatment and SCN infection. Molecular, biochemical, and genetic analyses showed that GmTINY binds to the Rhg1 ERELEE4 elements and regulates the expression of GmAATRhg1 and GmSNAP18, thereby enhancing SCN resistance. Additionally, overexpression of GmTINY induced several xyloglucan endotransglycosylase/hydrolase (XTH) genes, particularly GmXTH2, which is implicated in cell wall remodeling and restricts nematode development beyond the J2 stage. Together, these findings reveal an ETH-GmTINY signaling axis that regulates both Rhg1-mediated and cell wall-related resistance pathways, providing new insights for engineering durable SCN resistance in soybean.
Keywords: AP2/ERF transcription factor; GmTINY; Rhg1 locus; SCN; XTH; ethylene; soybean cyst nematode; xyloglucan endotransglycosylase/hydrolase.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Sugar Transporter GmSWEET20 Contributes to the Resistance of Soybean to Cyst Nematode.Phytopathology. 2025 Jul;115(7):881-890. doi: 10.1094/PHYTO-02-25-0059-R. Epub 2025 Jul 15. Phytopathology. 2025. PMID: 40262190
-
Genome-wide study and expression analysis of soybean ERF transcription factors and overexpression of GmERF205 enhances drought resistance in soybean.BMC Genomics. 2025 Aug 6;26(1):726. doi: 10.1186/s12864-025-11829-x. BMC Genomics. 2025. PMID: 40770289 Free PMC article.
-
GmLac55 Enhanced Soybean Resistance Against Soybean Cyst Nematodes Through Lignin Biosynthesis.Int J Mol Sci. 2025 Jun 30;26(13):6304. doi: 10.3390/ijms26136304. Int J Mol Sci. 2025. PMID: 40650081 Free PMC article.
-
Genome-wide association study and molecular marker development for resistance to soybean cyst nematode in soybean.Plant Genome. 2025 Sep;18(3):e70083. doi: 10.1002/tpg2.70083. Plant Genome. 2025. PMID: 40790875 Free PMC article.
-
Breeding for disease resistance in soybean: a global perspective.Theor Appl Genet. 2022 Nov;135(11):3773-3872. doi: 10.1007/s00122-022-04101-3. Epub 2022 Jul 5. Theor Appl Genet. 2022. PMID: 35790543 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

