Siamese neural network-enhanced electrocardiography can re-identify anonymized healthcare data
- PMID: 40395429
- PMCID: PMC12088719
- DOI: 10.1093/ehjdh/ztaf011
Siamese neural network-enhanced electrocardiography can re-identify anonymized healthcare data
Abstract
Aims: Many research databases with anonymized patient data contain electrocardiograms (ECGs) from which traditional identifiers have been removed. We evaluated the ability of artificial intelligence (AI) methods to determine the similarity between ECGs and assessed whether they have the potential to be misused to re-identify individuals from anonymized datasets.
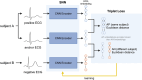
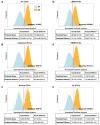
Methods and results: We utilized a convolutional Siamese neural network (SNN) architecture, which derives a Euclidean distance similarity metric between two input ECGs. A secondary care dataset of 864 283 ECGs (72 455 subjects) was used. Siamese neural network-electrocardiogram (SNN-ECG) achieves an accuracy of 91.68% when classifying between 2 689 124 same-subject pairs and 2 689 124 different-subject pairs. This performance increases to 93.61% and 95.97% in outpatient and normal ECG subsets. In a simulated 'motivated intruder' test, SNN-ECG can identify individuals from large datasets. In datasets of 100, 1000, 10 000, and 20 000 ECGs, where only one ECG is also from the reference individual, it achieves success rates of 79.2%, 62.6%, 45.0%, and 40.0%, respectively. If this was random, the success would be 1%, 0.1%, 0.01%, and 0.005%, respectively. Additional basic information, like subject sex or age-range, enhances performance further. We also found that, on the subject level, ECG pair similarity is clinically relevant; greater ECG dissimilarity associates with all-cause mortality [hazard ratio, 1.22 (1.21-1.23), P < 0.0001] and is additive to an AI-ECG model trained for mortality prediction.
Conclusion: Anonymized ECGs retain information that may facilitate subject re-identification, raising privacy and data protection concerns. However, SNN-ECG models also have positive uses and can enhance risk prediction of cardiovascular disease.
Keywords: Artificial intelligence; Continuous monitoring; Electrocardiogram; Identification; Siamese neural network.
© The Author(s) 2025. Published by Oxford University Press on behalf of the European Society of Cardiology.
Conflict of interest statement
Conflict of interest: J.W.W. and D.B.K. were previously on the advisory board for HeartcoR Solutions LLC. J.W.W. has received research support from Anumana. F.S.N. reports speaker fees from GE HealthCare and is on the advisory board for AstraZeneca. The remaining authors have no conflicts to declare.
Figures



Similar articles
-
Artificial intelligence-enhanced electrocardiography for the identification of a sex-related cardiovascular risk continuum: a retrospective cohort study.Lancet Digit Health. 2025 Mar;7(3):e184-e194. doi: 10.1016/j.landig.2024.12.003. Lancet Digit Health. 2025. PMID: 40015763
-
An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction.Lancet. 2019 Sep 7;394(10201):861-867. doi: 10.1016/S0140-6736(19)31721-0. Epub 2019 Aug 1. Lancet. 2019. PMID: 31378392
-
A comparison of artificial intelligence-enhanced electrocardiography approaches for the prediction of time to mortality using electrocardiogram images.Eur Heart J Digit Health. 2024 Nov 18;6(2):180-189. doi: 10.1093/ehjdh/ztae090. eCollection 2025 Mar. Eur Heart J Digit Health. 2024. PMID: 40110221 Free PMC article.
-
Artificial intelligence-enhanced electrocardiography for accurate diagnosis and management of cardiovascular diseases.J Electrocardiol. 2024 Mar-Apr;83:30-40. doi: 10.1016/j.jelectrocard.2024.01.006. Epub 2024 Jan 28. J Electrocardiol. 2024. PMID: 38301492 Review.
-
The Role of Machine Learning in the Detection of Cardiac Fibrosis in Electrocardiograms: Scoping Review.JMIR Cardio. 2024 Dec 30;8:e60697. doi: 10.2196/60697. JMIR Cardio. 2024. PMID: 39753213 Free PMC article.
Cited by
-
Computational modelling of biological systems now and then: revisiting tools and visions from the beginning of the century.Philos Trans A Math Phys Eng Sci. 2025 May 8;383(2296):20230384. doi: 10.1098/rsta.2023.0384. Epub 2025 May 8. Philos Trans A Math Phys Eng Sci. 2025. PMID: 40336283 Free PMC article. Review.
References
-
- Goldberger AL, Amaral LAN, Glass L, Hausdorff JM, Ivanov PCH, Mark RG, et al. PhysioBank, PhysioToolkit, and PhysioNet. Circulation 2000;101:E215–E220. - PubMed
-
- Mittelstadt BD, Floridi L. The Ethics of Big Data: Current and Foreseeable Issues in Biomedical Contexts. Cham: Springer; 2016. p. 445–480. - PubMed
LinkOut - more resources
Full Text Sources
