Mechanical force induced activation of adhesion G protein-coupled receptor
- PMID: 40395494
- PMCID: PMC12082320
- DOI: 10.1016/j.mbm.2024.100078
Mechanical force induced activation of adhesion G protein-coupled receptor
Abstract
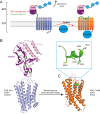
Among the various families of G protein-couple receptors (GPCR), the adhesion family of GPCRs is specialized by its expansive extracellular region, which facilitates the recruitment of various ligands. Previous hypothesis proposed that aGPCRs are activated by mechanical force, wherein a Stachel peptide is liberated from the GPCR autoproteolysis-inducing (GAIN) domain and subsequently binds to the transmembrane domain (7TM) upon activation. In this review, we summarize recent advancements in structural studies of aGPCRs, unveiling a conserved structural change of the Stachel peptide from the GAIN domain-embedded β-strand conformation to the 7TM-loaded α-helical conformation. Notably, using single-molecule studies, we directly observed the unfolding of GAIN domain and the release of Stachel peptide under physiological level of force, precisely supporting the mechanosensing mechanism for aGPCRs. We observed that the current complex structures of aGPCR adhesion domains with their respective ligands share a common pattern with the C-termini of each binding partner extending in opposite directions, suggesting a similar shearing stretch geometry for these aGPCRs to transmit the mechanical force generated in the circulating environment to the GAIN domain for its unfolding. Outstanding questions, including the relative orientations and interactions between 7TM and its preceding GAIN and adhesion domains of different aGPCRs, may require further structural and mechanical studies at the full-length receptor scale or cell-based level. Our analysis extends the current view of aGPCR structural organization and activation and offers valuable insights for the development of mechanosensor based on aGPCRs or discovery of mechanotherapy against aGPCRs.
Keywords: Adhesion GPCR; GAIN domain; Mechanosensing mechanism; Stachel peptide.
© 2024 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
Publication types
LinkOut - more resources
Full Text Sources

