Impaired K48-polyubiquitination downmodulates mouse norovirus propagation
- PMID: 40395509
- PMCID: PMC12089143
- DOI: 10.3389/fcimb.2025.1530166
Impaired K48-polyubiquitination downmodulates mouse norovirus propagation
Abstract
Introduction: Noroviruses are small non-enveloped, single stranded positive-sense RNA viruses that belong to the family Caliciviridae. They are highly contagious and resistant to multiple detergents and are the infectious agents in the majority of viral gastroenteritis in adults. Due to a lack of approved preventive or curative therapy options, intensive research effort is ongoing to better understand the pathogenesis mechanisms of noroviruses.
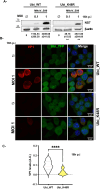
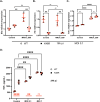
Methods: In this study, using the persistent murine norovirus S99 strain (MNoV_S99), we have investigated the role in the regulation of anti-noroviral responses of ubiquitination, a post-translational modification that covalently adds one or multiple ubiquitin molecules onto lysine residues of target proteins. To that end, we have first generated RAW264.7 cells overexpressing YFP-Ubiquitin_WT, _K29R, _K48R or_K63R constructs. All non-WT constructs encode a ubiquitin fusion protein with one lysine mutated into an arginine residue, thus preventing the formation of the K29-, K48- or K63-dependent polyubiquitin chains respectively.
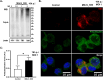
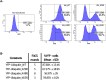
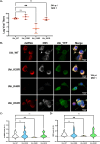
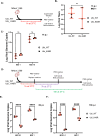
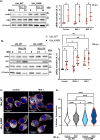
Results: Upon infection of these cells with MNoV_S99, we unexpectedly observed that only cells expressing the YFP-Ubiquitin_K48R protein showed a significantly impaired expression of several viral markers: NS5, NS7, VP1 and the replication intermediate dsRNA. Consequently, the number of viral genome copies or viral titers were also significantly decreased in the YFP-Ubiquitin_K48R cells compared to the YFP-Ubiquitin_WT cells. This negative regulation cannot be explained by perturbed viral entry, but rather a constitutive hypersecretion of the pro-inflammatory cytokine TNF and downstream upregulation of IκBα phosphorylation and the subsequent NF-κB nuclear translocation.
Conclusion: Overall, these consequences combined impose a non-permissive environment for MNoV_S99 replication and propagation.
Keywords: inflammation; innate immunity; macrophage; norovirus; signaling; ubiquitn.
Copyright © 2025 Wakeford, Werkmeister, Cayet, Poiret, Daniel, Mackenzie, Sirard, Lafont and Muharram.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures
Similar articles
-
CD300lf Conditional Knockout Mouse Reveals Strain-Specific Cellular Tropism of Murine Norovirus.J Virol. 2021 Jan 13;95(3):e01652-20. doi: 10.1128/JVI.01652-20. Print 2021 Jan 13. J Virol. 2021. PMID: 33177207 Free PMC article.
-
HOIL1 Is Essential for the Induction of Type I and III Interferons by MDA5 and Regulates Persistent Murine Norovirus Infection.J Virol. 2018 Nov 12;92(23):e01368-18. doi: 10.1128/JVI.01368-18. Print 2018 Dec 1. J Virol. 2018. PMID: 30209176 Free PMC article.
-
Identification of Antinorovirus Genes in Human Cells Using Genome-Wide CRISPR Activation Screening.J Virol. 2018 Dec 10;93(1):e01324-18. doi: 10.1128/JVI.01324-18. Print 2019 Jan 1. J Virol. 2018. PMID: 30305350 Free PMC article.
-
Regulation of HTLV-1 tax stability, cellular trafficking and NF-κB activation by the ubiquitin-proteasome pathway.Viruses. 2014 Oct 23;6(10):3925-43. doi: 10.3390/v6103925. Viruses. 2014. PMID: 25341660 Free PMC article. Review.
-
Lysine 63 polyubiquitination in immunotherapy and in cancer-promoting inflammation.Clin Cancer Res. 2009 Nov 15;15(22):6751-7. doi: 10.1158/1078-0432.CCR-09-1225. Epub 2009 Nov 3. Clin Cancer Res. 2009. PMID: 19887490 Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

