Autophagy in plants
- PMID: 40395529
- PMCID: PMC11864678
- DOI: 10.1080/27694127.2024.2395731
Autophagy in plants
Abstract
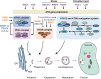
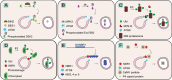
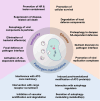
Autophagy is a process of cellular self-eating, which allows organisms to eliminate and recycle unwanted components and damaged organelles to maintain cellular homeostasis. It is an important process in the development of eukaryotic organisms. Autophagy plays a critical role in many physiological processes in plants such as nutrient remobilization, cell death, immunity, and abiotic stress responses. Autophagy thus represents an obvious target for generating resilient crops. During plant development, autophagy is also implicated in the differentiation and maturation of various cell types and plant organs, including root cap cells, tracheary elements, gametes, fruits and seeds. Here, we review our current understanding and recent advances of plant autophagy including insight into autophagy regulation and signaling as well as autophagosome membrane biogenesis. In addition, we describe how autophagy contributes to development, metabolism, biotic and abiotic stress tolerance and where the autophagic field is heading in terms of applied research for crop improvement.
Keywords: Plant autophagy; cargo receptors; crop improvement; development; endomembrane trafficking; immunity; metabolism; quality control; regulation and signalling; stress tolerance.
© 2024 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Figures


References
-
- Ding X, Zhang X, Otegui MS.. Plant autophagy: new flavors on the menu. Current opinion in plant biology. 2018. Dec;46:113–61. - PubMed
-
- van Doorn WG, Papini A.. Ultrastructure of autophagy in plant cells: a review. Autophagy. 2013. Dec;9(12):1922–1936. - PubMed
-
- Avin-Wittenberg T, Baluska F, Bozhkov PV, et al. Autophagy-related approaches for improving nutrient use efficiency and crop yield protection. J Exp Bot. 2018 Mar 14;69(6):1335–1353. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous
