Comprehensive Analysis of RNA Modifications Related Genes in the Diagnosis and Subtype Classification of Dilated Cardiomyopathy
- PMID: 40395552
- PMCID: PMC12089261
- DOI: 10.2147/JIR.S498496
Comprehensive Analysis of RNA Modifications Related Genes in the Diagnosis and Subtype Classification of Dilated Cardiomyopathy
Abstract
Background: RNA modifications are associated to various human diseases. However, the functions of RNA modification-related genes have yet to be thoroughly investigated in dilated cardiomyopathy (DCM). This study sought to conduct a comprehensive analysis of RNA modification-associated genes for the diagnosis and subtype classification of DCM.
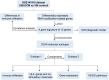
Methods: We collected DCM and control sample RNA modification-related genes from Gene Expression Omnibus (GEO) microarray datasets. Differential expression analysis was performed on these using the "Limma" package in R. Univariate logistic regression, and the LASSO algorithm were used to identify optimal genes for diagnostic model establishment. Furthermore, ConsensusClusterPlus was used to identify RNA modification-molecular subtypes. Lastly, the expression of the hub RNA modification-related genes and their connection to DCM were confirmed using the clinical samples and mouse models.
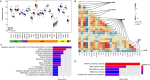
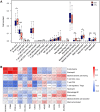
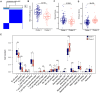
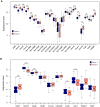
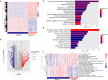
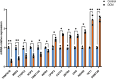
Results: Twenty-six RNA modification-related genes were identified as dysregulated in DCM, with strong connections noted among these genes. A diagnostic model based on 13 genes (TRMT61B, MBD2, YTHDC2, NOP2, TRMT10C, WDR4, CPSF2, CSTF3, ZBTB4, UNG, NSUN6, TET1, and DNMT3B) with an AUC of 0.980 predicted DCM well. Infiltrating plasma B cells, eosinophils, CD8 T cells, and regulatory T cells correlated strongly with TRMT61B, MBD2, YTHDC2, and CPSF2. Two RNA modification-molecular subtypes (clusters 1 and 2) were identified. Cluster 1 had greater RNA modification scores, lower immune ratings, and lower HLA-DRB1 and HLA-DPB1 expression than Cluster 2. Cluster 2 engaged metabolism-related pathways, while Cluster 1 activated renin-angiotensin system pathways.We further found a substantial link between lower cardiac function and up-regulation of TET1, DNMT3B, and down-regulation of MBD2, TRMT61B in the 13 hub RNA modification-related genes.
Conclusion: In conclusion, our RNA modification-related diagnostic model predicts DCM well. The discovery of two RNA modification-molecular subgroups and four key pivotal genes may assist stratify DCM patients by risk.
Keywords: 5-methylcytosine; N6-methyladenosine; RNA modification; dilated cardiomyopathy; immune infiltration.
© 2025 Xu et al.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this paper.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

