Retroviral adapters hijack the RNA helicase UPF1 in a CRM1/XPO1-dependent manner and reveal proviral roles of UPF1
- PMID: 40396490
- PMCID: PMC12093142
- DOI: 10.1093/nar/gkaf434
Retroviral adapters hijack the RNA helicase UPF1 in a CRM1/XPO1-dependent manner and reveal proviral roles of UPF1
Abstract
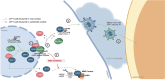
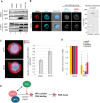
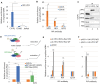
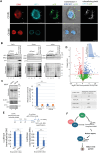
The hijacking of CRM1 export is an important step of the retroviral replication cycle. Here, we investigated the consequences of this hijacking for the host. During HTLV-1 infection, we identified that this hijacking by the viral protein Rex favours the association between CRM1 and the RNA helicase UPF1, leading to a decreased affinity of UPF1 for cellular RNA and its nuclear retention. As a consequence, we found that the nonsense-mediated mRNA decay (NMD), known to have an antiviral function, was inhibited. Corroborating these results, we described a similar process with Rev, the functional homolog of Rex from HIV-1. Unexpectedly, we also found that, for HTLV-1, this process is coupled with the specific loading of UPF1 onto vRNA, independently of NMD. In this latter context, UPF1 positively regulates several steps of the viral replication cycle, from the nuclear export of vRNA to the production of mature viral particles.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study, in the writing of the manuscript, or in the decision to publish the results.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

