The risk of pathogenicity and antibiotic resistance in deep-sea cold seep microorganisms
- PMID: 40396743
- PMCID: PMC12172429
- DOI: 10.1128/msystems.01571-24
The risk of pathogenicity and antibiotic resistance in deep-sea cold seep microorganisms
Abstract
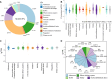
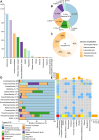
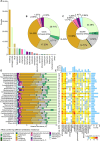
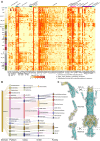
Deep-sea cold seeps host high microbial biomass and biodiversity that thrive on hydrocarbon and inorganic compound seepage, exhibiting diverse ecological functions and unique genetic resources. However, potential health risks from pathogenic or antibiotic-resistant microorganisms in these environments remain largely overlooked, especially during resource exploitation and laboratory research. Here, we analyzed 165 metagenomes and 33 metatranscriptomes from 16 global cold seep sites to investigate the diversity and distribution of virulence factors (VFs), antibiotic resistance genes (ARGs), and mobile genetic elements (MGEs). A total of 2,353 VFs are retrieved in 689 metagenome-assembled genomes (MAGs), primarily associated with indirect pathogenesis like adherence. In addition, cold seeps harbor nearly 100,000 ARGs, as important reservoirs, with high-risk ARGs (11.22%) presenting at low abundance. Compared to other environments, microorganisms in cold seeps exhibit substantial differences in VF and ARG counts, with potential horizontal gene transfer facilitating their spread. These virulome and resistome profiles provide valuable insights into the evolutionary and ecological implications of pathogenicity and antibiotic resistance in extreme deep-sea ecosystems. Collectively, these results indicate that cold seep sediments pose minimal public health risks, shedding light on environmental safety in deep-sea resource exploitation and research.
Importance: In the "One Health" era, understanding pathogenicity and antibiotic resistance in vast and largely unexplored regions like deep-sea cold seeps is critical for assessing public health risks. These environments serve as critical reservoirs where resistant and virulent bacteria can persist, adapt, and undergo genetic evolution. The increasing scope of human activities, such as deep-sea mining, is disrupting these previously isolated ecosystems, heightening the potential for microbial exchange between deep-sea communities and human or animal populations. This interaction poses a significant risk for the dissemination of resistance and virulence genes, with potential consequences for global public health and ecosystem stability. This study offers the first comprehensive analysis of virulome, resistome, and mobilome profiles in cold seep microbial communities. While cold seeps act as reservoirs for diverse ARGs, high-risk ARGs are rare, and most VFs were low risk that contribute to ecological functions. These results provide a reference for monitoring the spread of pathogenicity and resistance in extreme ecosystems, informing environmental safety assessments during deep-sea resource exploitation.
Keywords: antibiotic resistance genes; cold seeps; microbiome; virulence factors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Miller KA, Thompson KF, Johnston P, Santillo D. 2018. An overview of seabed mining including the current state of development, environmental impacts, and knowledge gaps. Front Mar Sci 4. doi: 10.3389/fmars.2017.00418 - DOI
MeSH terms
Substances
Grants and funding
- No. 3502Z202373076/Natural Science Foundation of Xiamen, China
- No. 2023J06042/Natural Science Foundation of Fujian Province
- No. 42376115, No. 92351304/National Natural Science Foundation of China
- No. 2022025, No. 2023022/Scientific Research Foundation of Third Institute of Oceanography, MNR
- No. 2021R51008/Zhejiang Provincal High-level Talent Special Support Plan
LinkOut - more resources
Full Text Sources
Medical

