Cullin 3-mediated ubiquitination restricts enterovirus D68 replication and is counteracted by viral protease 3C
- PMID: 40396757
- PMCID: PMC12172421
- DOI: 10.1128/jvi.00354-25
Cullin 3-mediated ubiquitination restricts enterovirus D68 replication and is counteracted by viral protease 3C
Abstract
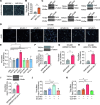
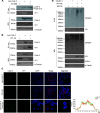
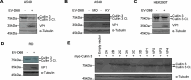
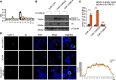
Enterovirus D68 (EV-D68) has emerged as a significant threat to public health because of its association with respiratory illnesses and neurological complications, including acute flaccid myelitis. However, the molecular mechanisms underlying EV-D68 replication and pathogenesis remain unclear. Here, we revealed a novel interaction between EV-D68 and the host Cullin-RING E3 ligase system, specifically Cullin 3, which was reported to restrict viral replication. We initially demonstrated that proteasome inhibition enhanced EV-D68 replication, suggesting an important role for the ubiquitin-proteasome system in viral restriction. Cullin 3 was further identified as a key factor that inhibits EV-D68 replication, and the downregulation of its expression increased viral titers. Mechanistically, Cullin 3 was observed to target the viral capsid protein VP1 for ubiquitination and degradation. However, EV-D68 was determined to utilize its protease 3C to cleave Cullin 3 at the Q681 residue, thereby inhibiting E3 ligase activity and facilitating resistance to Cullin 3-mediated VP1 degradation. This study uncovered a host-virus arms race, wherein the ubiquitin-proteasome system of the host actively targets viral proteins for degradation, and viral proteases counteract this defense mechanism. Accordingly, these findings could lead to more effective antiviral treatments.
Importance: The ubiquitin-proteasome system (UPS) is a critical cellular pathway involved in the regulation of protein stability and has been implicated in the regulation of viral infections. However, its role in EV-D68 infection has not been extensively explored. Our study proves that the host UPS, through the scaffold protein Cullin 3, can restrict EV-D68 replication, representing a previously unrecognized antiviral mechanism. Furthermore, we describe a viral strategy used to evade this host defense mechanism comprising Cullin 3 cleavage, which has broad implications for understanding virus-host interactions and could inform the development of novel therapeutic strategies against EV-D68 and other enteroviruses.
Keywords: Cullin 3; enterovirus D68; protein cleavage; protein degradation; ubiquitination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Enteroviral 3C protease cleaves N4BP1 to impair the host inflammatory response.J Virol. 2025 Jan 31;99(1):e0175824. doi: 10.1128/jvi.01758-24. Epub 2024 Dec 10. J Virol. 2025. PMID: 39655957 Free PMC article.
-
The use of sialic acids as attachment factors is a common feature of Enterovirus-D species.J Virol. 2025 Jun 17;99(6):e0042925. doi: 10.1128/jvi.00429-25. Epub 2025 May 13. J Virol. 2025. PMID: 40358210 Free PMC article.
-
Enhanced genomic surveillance of enteroviruses reveals a surge in enterovirus D68 cases, the Johns Hopkins health system, Maryland, 2024.J Clin Microbiol. 2025 Jul 9;63(7):e0046925. doi: 10.1128/jcm.00469-25. Epub 2025 Jun 10. J Clin Microbiol. 2025. PMID: 40492725 Free PMC article.
-
Global emergence of enterovirus D68: a systematic review.Lancet Infect Dis. 2016 May;16(5):e64-e75. doi: 10.1016/S1473-3099(15)00543-5. Epub 2016 Feb 24. Lancet Infect Dis. 2016. PMID: 26929196
-
Ubiquitin proteasome system (UPS): a crucial determinant of the epigenetic landscape in cancer.Epigenomics. 2025 Jun;17(9):625-644. doi: 10.1080/17501911.2025.2501524. Epub 2025 May 8. Epigenomics. 2025. PMID: 40337853 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

