Structure of the microtubule-anchoring factor NEDD1 bound to the γ-tubulin ring complex
- PMID: 40396914
- PMCID: PMC12094035
- DOI: 10.1083/jcb.202410206
Structure of the microtubule-anchoring factor NEDD1 bound to the γ-tubulin ring complex
Abstract
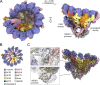
The γ-tubulin ring complex (γ-TuRC) is an essential multiprotein assembly that provides a template for microtubule nucleation. The γ-TuRC is recruited to microtubule-organizing centers (MTOCs) by the evolutionarily conserved attachment factor NEDD1. However, the structural basis of the NEDD1-γ-TuRC interaction is not known. Here, we report cryo-EM structures of NEDD1 bound to the human γ-TuRC in the absence or presence of the activating factor CDK5RAP2. We found that the C-terminus of NEDD1 forms a tetrameric α-helical assembly that contacts the lumen of the γ-TuRC cone and orients its microtubule-binding domain away from the complex. The structure of the γ-TuRC simultaneously bound to NEDD1 and CDK5RAP2 reveals that both factors can associate with the "open" conformation of the complex. Our results show that NEDD1 does not induce substantial conformational changes in the γ-TuRC but suggest that anchoring of γ-TuRC-capped microtubules by NEDD1 would be structurally compatible with the significant conformational changes experienced by the γ-TuRC during microtubule nucleation.
© 2025 Muñoz-Hernández et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures







Update of
-
Structure of the microtubule anchoring factor NEDD1 bound to the γ-tubulin ring complex.bioRxiv [Preprint]. 2024 Nov 5:2024.11.05.622067. doi: 10.1101/2024.11.05.622067. bioRxiv. 2024. Update in: J Cell Biol. 2025 Aug 4;224(8):e202410206. doi: 10.1083/jcb.202410206. PMID: 39574704 Free PMC article. Updated. Preprint.
Similar articles
-
Structure of the microtubule anchoring factor NEDD1 bound to the γ-tubulin ring complex.bioRxiv [Preprint]. 2024 Nov 5:2024.11.05.622067. doi: 10.1101/2024.11.05.622067. bioRxiv. 2024. Update in: J Cell Biol. 2025 Aug 4;224(8):e202410206. doi: 10.1083/jcb.202410206. PMID: 39574704 Free PMC article. Updated. Preprint.
-
Structure of the γ-tubulin ring complex-capped microtubule.Nat Struct Mol Biol. 2024 Jul;31(7):1124-1133. doi: 10.1038/s41594-024-01264-z. Epub 2024 Apr 12. Nat Struct Mol Biol. 2024. PMID: 38609661 Free PMC article.
-
Microtubule nucleation: How the NEDD1:MZT1:GCP3 trio captures the γ-TuRC.J Cell Biol. 2025 Aug 4;224(8):e202506019. doi: 10.1083/jcb.202506019. Epub 2025 Jul 15. J Cell Biol. 2025. PMID: 40663060
-
Microtubule nucleation: The waltz between γ-tubulin ring complex and associated proteins.Curr Opin Cell Biol. 2021 Feb;68:124-131. doi: 10.1016/j.ceb.2020.10.004. Epub 2020 Nov 12. Curr Opin Cell Biol. 2021. PMID: 33190097 Review.
-
The structure of the γ-TuRC at the microtubule minus end - not just one solution.Bioessays. 2024 Sep;46(9):e2400117. doi: 10.1002/bies.202400117. Epub 2024 Jul 23. Bioessays. 2024. PMID: 39044599 Review.
Cited by
-
Cryo-EM structures of the Plant Augmin reveal its intertwined coiled-coil assembly, antiparallel dimerization and NEDD1 binding mechanisms.bioRxiv [Preprint]. 2025 Feb 27:2025.02.25.640204. doi: 10.1101/2025.02.25.640204. bioRxiv. 2025. PMID: 40034650 Free PMC article. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

