ADP-ribosylation of NuMA promotes DNA single-strand break repair and transcription
- PMID: 40397572
- PMCID: PMC12187637
- DOI: 10.1016/j.celrep.2025.115737
ADP-ribosylation of NuMA promotes DNA single-strand break repair and transcription
Abstract
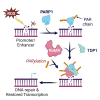
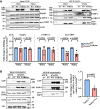
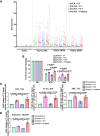
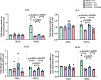
Single-strand breaks (SSBs) are prevalent DNA lesions implicated in genome instability. The nuclear mitotic apparatus protein (NuMA) has been reported to promote SSB repair (SSBR) and regulate transcription following oxidative stress. ADP-ribosylation, an important post-translational modification, regulates several processes, including chromatin remodeling, transcription, and DNA repair. To investigate its role in NuMA-dependent functions, we generated an ADP-ribosylation-deficient NuMA construct and report that NuMA ADP-ribosylation is required for its interaction with tyrosyl DNA phosphodiesterase 1 (TDP1), an SSBR player. Cells expressing ADP-ribosylation-deficient NuMA exhibit delayed SSBR kinetics following oxidative stress and reduced repair at promoter and enhancer regions, consistent with a role of NuMA in protecting non-coding regulatory regions from DNA damage. Furthermore, the expression of NuMA-regulated genes following oxidative stress requires ADP-ribosylation. Our findings demonstrate that ADP-ribosylation of NuMA promotes SSBR and transcription following oxidative stress, underscoring the importance of ADP-ribosylation in modulating DNA repair and gene expression.
Keywords: ADP-ribosylation; CP: Molecular biology; DDR; DNA damage response; DNA repair; IEGs; NuMA; brain health; cancer; dementia; gene regulatory elements; immediate early genes; oxidative DNA damage; oxidative stress; single-strand DNA breaks; transcription.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

